The DNA sequencing picture that emerges today shows the central Honshu people of Japan to be genetically just a little closer to Sino-Tibetan and Han Chinese (from the Jiangsu region who were possibly rice-farming immigrants during the Yayoi era) evidenced by the specific genetic Y markers found in Japanese today (ie O3a5, O3a and O1), in their mix than to modern-day Koreans whose ancestors contributed significantly to the Japanese gene pool probably during Koguryo and Paekche migrations into Japan of the Kofun era to Asuka eras.
One surprising point that emerges from a look at both mtDNA and Y Haplogroups charts, is that the Koreans show an even closer genetic affinity to Okinawans (and therefore to the Jomon stock) than mainland Honshu Japanese do themselves…comprising 17.4% of their DNA sequence compared to the Japanese 16.1% of their DNA sequence.
Satoshi Yamaguchi’s studies
In recent years, more archaeological and genetic evidence have been found in both eastern China and western Japan to lend credibility to this argument. Between 1996 and 1999, a team led by Satoshi Yamaguchi, a researcher at Japan’s National Science Museum, compared Yayoi remains found in Japan’s Yamaguchi and Fukuoka prefectures with those from early Han Dynasty (202 BC-8) in China’s coastal Jiangsu province, and found many similarities between the skulls and limbs of Yayoi people and the Jiangsu remains. Two Jiangsu skulls showed spots where the front teeth had been pulled, a practice common in Japan in the Yayoi and preceding Jōmon period. The genetic samples from three of the 36 Jiangsu skeletons also matched part of the DNA base arrangements of samples from the Yayoi remains. Surprisingly, Japanese also display the highest frequency of haplogroup O3a5, which is a Han Chinese and Sino-Tibetan specific O3 branch.
Japanese Haplogroup O3a5 (O3e) 10/47= 23%
This frequency is about 5% higher than the frequency of O3a5 among Manchus, Koreans and other Northeast Asians.
For North Koreans, the frequency of O3a5 is even lower than some Tungusic populations. Overall, the Koreanic haplogroup O3 were the least influenced by Sinitic populations.
Whereas pure haplogroup C3 (M217-no subclade) was observed at a high frequency among Tungusic (20%) and Koreanic (16%) populations. The frequency of haplogroup C3 among Japanese was only 1%. This means that Japanese origins were not as prominently from Siberia as was commonly thought, since Japanese bear more of C1, whereas C3 is found only in northern populations of Japan.
Haplogroup D was observed among Japanese (25%) and Tibetans (40%); it was also observed among Han Chinese, Mongolians and Koreans.
The DNA sequence SNP study done by Japanese researchers in 2005 (the biggest contributor of DNA of each East Asian people is bolded) showed the following results:
Korean DNA sequence is made up of:
40.6% Uniquely Korean
21.9% Chinese
1.6% Ainu
17.4% Okinawan
18.5% Unidentified
Japanese DNA sequence is made up of:
4.8% Uniquely Japanese
24.2% Korean
25.8% Chinese
8.1% Ainu
16.1% Okinawan
21% Unidentified
Chinese DNA sequence is made up of:
60.6% Uniquely Chinese
1.5% Japanese
10.6% Korean
1.5% Ainu
10.6% Okinawan
15.2% Unidentified
The biggest components in Japanese are Chinese, Korean, Okinawan.
A closer look at the Chinese gene pool
The shared Chinese gene pool between Japanese and Koreans is thought to be formed of Dong-Yi stock (originating from China’s Shandong peninsula) that later formed the Puyo peoples of the Paekche kingdom and Koguyro kingdoms of the Korean peninsula.
mtDNA haplogroup A which is widespread in Asia today occurs at levels below 10%, but reaches higher concentrations in some parts of China, Korea and Japan.
“Some ethnic Chinese populations, such as the Dong and the Yi, carry haplogroup A at levels as high as 30%. One branch of the haplogroup, A4, reaches levels of more than 15% among mitochondrial DNA samples collected in the city of Wuhan in central China. In the Spittoon… Ancient China’s famous Terracotta Army was constructed by men bearing haplogroup A. Check the Spittoon to learn more about these ancient builders.” — Source: An Introduction to Haplogroups: An Interactive Activity Activity developed by Meredith T. Knight at Tufts University
Ancient mtDNA in Siberia Haplogroup A was widespread in Siberia in ancient times. One study of skeletal remains discovered near Siberia's Lake Baikal estimated the haplogroup was present in 13-26% of the region's population 7,000 years ago, and is almost exclusively among the Chukchi and the Yupik, two small indigenous groups from northeastern Siberia.
M7, a widespread haplogroup found in China, Japan, Southeast Asia and the Pacific Islands. [M8, a widespread haplogroup in central and eastern Asia that eventually sent an offshoot to the Americas. M9, which appears to have arisen in Tibet.] While Haplogroup M is widespread throughout South and East Asia, it originates from the Indian sub-continent where it is more diverse on there than anywhere else in the world.
Overall, Japanese are closest to Tibetans and Han Chinese, but only marginally more so than to the Koreans.
The Ainu who are widely considered to be of the “old” proto-Mongoloid stock closely related to the Tibetan Buryat and Yakut peoples, and descended from the Jomon people who lived in the Tohoku area until they were later pushed northwards into Hokkaido, afterwhich they resided around the Sea of Okhotsk, mainly Hokkaido, Sakhalin, Kuril Islands and the tip of Kamchatka. However, the DNA sequences show that Ainu are actually more remotely distanced from the Jomon than is commonly believed, as they were influenced by Siberians (as with Koreans). Evidence is the haplogroup C3 (no subclade) occurs at moderately high frequencies among these populations.
The above mtDNA studies relate to maternal line gene flow, the following Y chromosome study of male-mediated gene flow shows a slightly different picture but the sharing of the common haplotypes still reveal strong affiliations of both Japanese and Koreans to the Chinese and of the Japanese to the Koreans:
Population studies of genetic markers such as HLA variation and mitochondrial DNA have been used to understand human origins, demographic and migration history. Recently, diversity on the nonrecombining portion of the Y chromosome (NRY) has been applied to the study of human history. Since NRY is passed from father to son without recombination, polymorphisms in this region are valuable for investigating male-mediated gene flow and for complementing maternally based studies of mtDNA. Haplotypes constructed from Y-chromosome markers were used to trace the paternal origins of Korean. By using 38 Y chromosome single nucleotide polymorphism markers, the genetic structure of 195 Korean males was analyzed.
The Korean males were characterized by a diverse set of 4 haplogroups (Groups IV, V, VII, X) and 14 haplotypes that were also present in Chinese.
The most frequent haplogroup in Korean was Group VII (82.6%). It was also the most frequent haplogroup in Chinese (95%) as well as in Japanese (45%). The frequencies of the haplogroups V, IV, and X were 15.4%, 1%, and 1%, respectively. The second most frequent haplogroup V in Korean was not present in Chinese, but its frequency was similar in Japanese.
Source of study: Sunghee Hong, Seong-Gene Lee, Yongsook Yoon, Kyuyoung Song /University of Ulsan College of Medicine, 388-1 Poongnap-dong, Songpa-ku, Seoul, Korea
Finally, the Y haplogroup chart below shows the relations between the various groups of Asia and their varying degrees of affinity or remoteness to each other.
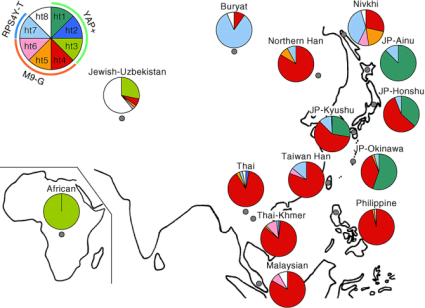
Tajima: Frequency distributions for for 8 Y-chromosome haplotypes for 14 global populations(Ainu, Honshu and Okinawan Japanese relationships seen)
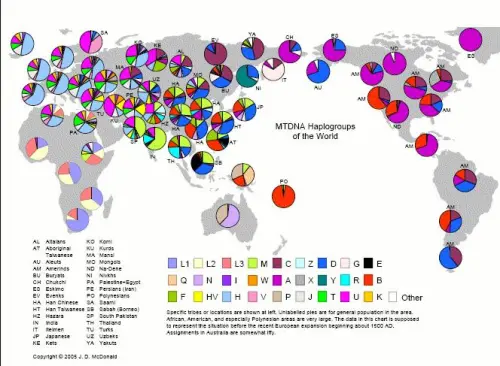
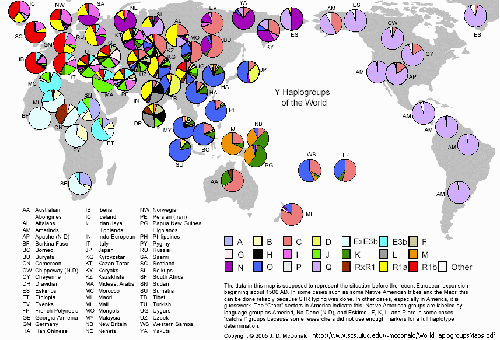
Y Haplogroups of the world
Another study published in 2005 on 81 sets of Y chromosomes of six populations across Japan showed:
– The Japanese have at least two very deep pre-Yayoi ancestral Y chromosome lineages (D-P37.1 and C-M8) that descend from Paleolithic founders who had diverged from the mainland and that were then isolated from those populations on the mainland for a very long time. Scientists thought these D lineages to mean the Jomon populations in Japan once upon a time the same ancestors as Tibetans from central Asia who are found with the highest frequency of continental D lineages is found in central Asia. Scientists hypothesized that the area between Tibet and the Altai Mountains in northwestern China is the most likely geographic source of Paleolithic Japanese founding Y chromosomes. ( Historical records suggest that Tibetan populations were derived from ancient tribes of northwestern China that subsequently moved to the south and mixed with the southern natives in the last 3,000 years.) A separate recent mtDNA study on the Haplogroup M12 – the mitochondrial component of Japanese genes, the counterpart of Y chromosome D lineage – also confirmed the direct connections of Japanese haplotypes with Tibet. This rare haplogroup is possessed only by mainland Japanese, Koreans, and Tibetans, with the highest frequency and diversity in Tibet. These Paleolithic ancestors were thought to have migrated into Japan sometime around 20,000 years ago.
Conclusion
In a nutshell, what we can evince and conclude from all the DNA data that has been presented is that the Japanese people are a people with mixed diverse origins, formed from many waves of migrations from various locations in the remote past as well as in the more recent past.
References:
Tajima, Atsushi, Studies on the geographic distribution of the human Y DNA variation
Dual origins of the Japanese: common ground for hunter-gatherer and farmer Y chromosomes [full document here]
Genetic structure of the Japanese people and the formation of the Ainu population by Ken-ichi Shinoda (National Museum of Science and Nature, Tokyo)
Watch the documentary “Japanese Roots” at Youtube.
Tateno, Yoshio et al., Divergence of East Asians and Europeans Estimated Using Male- and Female-Specific Genetic Markers
World Haplogroup Maps Source: J.D. McDonald
Origins of the Jomon, connections with the Asian continent
Y Chromosomal DNA and the Peopling of Japan by Michael F. Hammer and Satoshi Horai
An Introduction to Haplogroups: An Interactive Activity Activity developed by Meredith T. Knight at Tufts University
B. Mohan Reddy et al., “Austro-Asiatic Tribes of Northeast India Provide Hitherto Missing Genetic Link between South and Southeast Asia” on the origin of O-M95 in northeast India
Longli Kang et al., Y chromosome O3 haplogroup diversity in Sino-Tibetan populations reveals two migration routes into the eastern Himalayas
Stoneking and Delfin, The human genetic history of East Asia: Weaving a complex tapestry

Thank you for this information. Too bad the koreans, chinese and japanese are still fighting so much. We are share some common genes. NO wonder we eat food that are so alike too.
so if you dont have ht1, then you are not japanese? where do i get tested?
i read this article that said a japanese doctor from the university of pensylvania carried out a research among japanese, koreans and tibetans to find out the origin of the japanese.it said he found that the japanese peoples were heavily influenced by the yayoi peoples dna.the yayoi peoples were from the korean peninsula and they had found a genetic marker among japanese and korean males that originated from the yayoi peoples. this genetic marker was also found at high frequencies among tibetan males.the researchers then concluded that this marker was brought to the korean peninsula by the ancestors of the yayoi peoples and also to tibet by the ancestors of the tibetans.
The koreans have like 20% chinese dna and about 21% japanese dna according to several researchers. KOreans have only like 40% unqiue korean dna and the remaining dna cannot be identify. DNA is muating faster than ever. Scientist said think it use to take like 20 to 30 thousands of years for genes to mutate, but science in the pats 10 years fund dna is mutating much faster than ever. Scientist better hurry it up before dna is untraceable. Human kind adapt to their climite and environment. We are came from east africia. We mutated according to the path we took. I don’t know why people keep fight who is better. It is so ridiculous!!!!!!!!! We are are human. Until outerspace aliens invade us in the future, then us the so call human race will see us as all the same.
” The koreans have like 20% chinese dna and about 21% japanese dna according to several researchers. ”
May I ask where you got this info from?
This data is based on a number of sources, one of then a 2003 Japanese DNA study mentioned here…
http://www.rjkoehler.com/2007/08/22/korea-a-nation-of-immigrants/
But much more higher resolution and recent study puts the figure at 30% for mtDNA and over 70% for YDNA see
Citation: Jin H-J, Tyler-Smith C, Kim W (2009) The Peopling of Korea Revealed by Analyses of Mitochondrial DNA and Y-Chromosomal Markers. PLoS ONE 4(1): e4210. doi:10.1371/journal.pone.0004210
http://www.plosone.org/article/info%3Adoi%2F10.1371%2Fjournal.pone.0004210
The Korean Peninsula is located to the north of the Yellow and Yangtze Rivers of China, and bounded to the northeast by Russia. Therefore, the Koreans are geographically a northeast Asian group. Anthropological and archeological evidence suggests that the early Korean population was related to Mongolian ethnic groups who inhabited the general area of the Altai Mountains and Lake Baikal regions of southeast Siberia [7]. According to Korea’s founding myths, the Ancient Chosun (the first state-level society of Korea) was established around 2,333 BC in the region of southern Manchuria but later moved into the Pyongyang area of northwest Korea. In addition, archeological evidence reveals that rice cultivation had spread to most parts of the Korean Peninsula by around 1,000–2,000 BC, introduced from the Yellow River and/or Yangtze River basin in China [8].
Studies of classical genetic markers showed that Koreans tend to have a close genetic affinity with Mongolians among East Asians [9]–[11]. In contrast, recent surveys of Y-chromosomal DNA variation revealed that the Korean population contained lineages typical of both southern and northern East Asian populations [6], [12], [13]. The Koreans appeared to have affinities with Manchurians, Yunnan-Chinese from southern China, and Vietnamese [13].
…Other Siberian and Mongolian-prevalent haplogroups from the C, Y and Z lineages make up less than 4% of the Korean mtDNA pool. Haplogroups A5a and Y2 are found almost exclusively in Korea but were present at extremely low frequencies. In total, these northern haplogroups account for ~60% of the mtDNA gene pool of the Koreans. In addition, southeast Asian-prevalent mtDNA lineages of (sub)haplogroups B (14.6%), M7 (10.3%), and F (9.7) are also found at moderate frequencies in the Korean population (Table 2). These findings suggest that more than 30% of the Korean mtDNA pool is attributable to maternal lineages with a more southern origin. We also found the haplogroup M7a1 exclusively in the Korean population. This result is consistent with previous reports that haplogroup M7a is restricted to Japan and south Korea [18], [20]. Thus, the distribution pattern of mtDNA haplogroups leads us to consider that the peopling of Korea is likely to have involved multiple sources.
Our study documents the genetic relationships of the Koreans with their neighboring populations in unprecedented detail. Two major findings emerge. First, the Koreans are overall more similar to northeast Asians than to southeast Asians. This conclusion would be expected from the general correlation between genetic variation and geography observed for human populations, and is supported here by an examination of individual mtDNA haplogroups (Table 2), genetic distances between populations derived from mtDNA or Y-chromosomal data (Figure 2), and the apportionment of genetic diversity between different groups of populations (Table 4). Second, the conclusions from mtDNA and Y-chromosomal analyses differ. Sex-biased admixture is common in human expansions such as that of Bantu-speaking farmers in Africa[58], the spread of the Han ethnic group in China [59] or the post-Columbian peopling of the Americas [60]. The effects in Korea are more subtle, but show a larger male than female contribution from southern East Asia to the population of Korea, most clearly revealed by the admixture estimates, where a 35% contribution from the south was estimated for mtDNA, compared with a 83% contribution for the Y chromosome (Table 5).
Table 5. Admixture estimates of Northeast Asians and Southeast Asians in Korean populations.
doi:10.1371/journal.pone.0004210.t005
The predominant genetic relationship with northern East Asians is consistent with other lines of evidence. Xue et al. [31] reported that the northern East Asian populations started to expand in number before the last glacial maximum at 21-18 KYA, while the southern populations all started to expand after it, but then grew faster, and they suggested that the northern populations expanded earlier because they could exploit the abundant megafauna of the “Mammoth Steppe,” while the southern populations could increase in number only when a warmer and more stable climate led to more plentiful plant resources such as tubers. By this criterion, the Koreans, expanding at about 30 KYA [31] also resemble other northern populations. Historical evidence suggests that the Ancient Chosun, the first state-level society, was established in the region of southern Manchuria and later moved into the Pyongyang area of the northwestern Korean Peninsula. Based on archeological and anthropological data, the early Korean population possibly had an origin in the northern regions of the Altai-Sayan and Baikal regions of Southeast Siberia [7], [8], [61].
What could be the origin of the male-biased southern contribution to Korean gene pool illustrated, for example, by haplogroups O-M122 (42.2%) and O-SRY465 (20.1%) [29]. Recent molecular genetic analyses and the geographical distribution of haplogroup O-M122 lineages, found widely throughout East Asia at high frequencies (especially in southern populations and China), have suggested a link between these Y-chromosome expansions and the spread of rice agriculture in East Asia [62]–[64]. In general, Y-chromosomes might be spread via a process of demic diffusion during the early agricultural expansion period [65], [66]. If this interpretation were substantiated, the spatial pattern of Y-haplogroup O would imply a genetic contribution to Korea through the spread of male-mediated agriculture. Large-scale genetic analyses thus begin to reveal some of the complexities of the peopling of Korea, and further studies of individual autosomal loci or genomewide genotyping and sequencing are expected to further insights…
“The koreans have like 20% chinese dna and about 21% japanese dna…”
I also wanted to ask, is this speaking about the whole Korean race that have these percentages of “chinese” and “japanese” DNA?
In other words, does 100% of Korea’s people have this or is it saying 50% of Korea’s population have this?
Does this make sense?
Thanks in advance.
Once joined a team like Yankees, DNA doesn’t matter, only hit rate, isn’t this true boys? So, it is here in Japan.
These DNA tests aren’t fully credible. I mean how we can know the basis of classification of similar or different. Are these classification really admitted internationally by all countries with no objections? This doesn’t include the relation of Hebrew, middle easterns and other Asians. Why do you just spot on these 3 countries. There are Mongol, Khazafstan, Indonesia etc in Asia. And our all ancestors were supposely born in Africa about 60,000 years ago. Biblical perspective is a shit compared to Science figure.
There are several problems. One of the most obvious problems is ascertaining timing of the branching of or origin of genotypes – one study puts it this way: “It is difficult to accurately date the ancient human migrations (or mutations), because of the errors inherently involved in estimating both the effective population size of the males and the mutation rate. However, our knowledge of morphology and archaeology can help us to narrow the estimated age range”. DNA science is still a relatively new field so that the information is only as good as the number of studies that have been done. This is not the same as saying that the data is not credible. Interpretation of that data is however, obviously only as good as the amount of data or studies that have been done on a specific topic. Central Asia and Mongolia were some of the regions that lacked DNA studies the most … until recently. The study of Y or mtDNA haplogroups or types are highly informative however, they provide very specific signals of the existence of a specific people within a population. You can track lineages of people within populations to reasonable accuracy, on account of the genetic mutations and know which ones came before which ones. As more and more genetic data and studies are published, we are beginning to get a clarifying picture of the migratory trails various lineages have left all over the world. The race has been on to establish the earliest human settlement into E. Asia, and the sequence of settler arrivals – earlier theories posited arrivals from the Eurasian heartland as the earliest , recent ones say SEA-Austronesian ones by way of S. Asia were earliest, others suggest out of Island SEA. The jury is still out on the issue. Obviously, since the regions yet to be studied in depth are East Asia and Central Asia… the picture still requires much clarifying something which has to be done by comparing data from different types of studies, Y or mtDNA/autosomal/HLA antigens/ virus types/ other disease genetic data and with archaeological and anthropological data. The picture of migratory trails and lineages that is emerging is extremely complex, but it is none the less emerging and clarifying. “Why do you just spot on 3 countries?” Good question. Partly for the lack of page space. But really because it’s an ongoing analysis. I’m currently working on the possible trails from the Altai-Baikal-Mongolia region, as well as the R1b1b trails as well as the J hg trails to and from the Near East/ME as well as the Indo-Aryan Indian subcontinent. But a great many new and exciting studies have come onstream from these areas – as well as on E. Asia … only very recently, particularly in 2010 and this year. It takes time to digest these, and draw inferences for a macro-viewpoint.
Let’s face it. People always move around, so it isn’t surprising to me that koreans have chinese and japanese DNA and vise versa. Japanese have korean and chinese dna, but What about chinese dna. I think they also have some variation of Japanese in northern or northeast china. My believe is migration out of africa came to populate CHina, Korea, Japan from east to west, so that explain why Japanese and korean male carry a tibetian gene. Also tibetians migrated up to the high plateau less than 3,000 years ago . TIbetian share gene pool with the ethnic chinese population except for a few gene that are different due to adaptation to the high altitude.
Damn people should put aside their national pride. I strongly feel that we mongoloid people are related and traveled the same path out of africa. Some went to central asia, north asia, northeast asia, east asia, and then some migrated into southeast asia like vietnam. Southeast asians are either pure australoid people are they are a mixture of australoid and mongoloid race. I would like to find out more in my life about the the mongoloid asians and how they are related to each other. I like them all. I don’t know why the hate. We are all people. We should judge by indivuality and not by ethnicity or the race of the person. No race or ethnicity is better than the other race or ethnicity. I encountered too much hate while watch youtube.
The confusion of most of the internet talk results when people forget that what’s deemed Chinese or Japanese or Korean have to do with statehood concepts and boundaries which appeared only around 2,000 years ago give or take. Beyond that, the picture is of nomadic tribal groups, chiefdoms, migratory groups whether farmers or pastoral nomads, and further back in time, hunter-gatherers. For example, a 2009 study on Korean populations showed “recent findings from Y chromosome studies showed that the Korean population contains lineages from both southern and northern parts of East Asia.” and that “haplogroup O-M122 lineages, found widely throughout East Asia at high frequencies (especially in southern populations and China), have suggested a link between these Y-chromosome expansions and the spread of rice agriculture in East Asia …archaeological evidence reveals that rice cultivation had spread to most parts of the Korean Peninsula by around 1,000–2,000 BC, introduced from the Yellow River and/or Yangtze River basin in China …northern haplogroups account for ∼60% of the mtDNA gene pool of the Koreans. In addition, southeast Asian-prevalent mtDNA lineages of (sub)haplogroups B (14.6%), M7 (10.3%), and F (9.7) are also found at moderate frequencies in the Korean population. These findings suggest that more than 30% of the Korean mtDNA pool is attributable to maternal lineages with a more southern origin. We also found the haplogroup M7a1 exclusively in the Korean population. This result is consistent with previous reports that haplogroup M7a is restricted to Japan and south Korea. Thus, the distribution pattern of mtDNA haplogroups leads us to consider that the peopling of Korea is likely to have involved multiple sources.” Modern-day East Asian populations (China, Korea and Japan) are like a combination of layered and marbled cake with visible threads of differing ingredients as well as admixtures as well. Many new studies are throwing light on these threads that run from the different migratory lines: Large-scale mtDNA screening reveals a surprising matrilineal complexity in east Asia and its implications to the peopling of the region; The emerging limbs and twigs of the East Asian mtDNA tree; The human genetic history of East Asia: weaving a complex tapestry. What we are needing to build a picture of peopling origins is a higher resolution scrutiny of the finer twigs of the branching lines…
This 2010 study Global distribution of Y-chromosome haplogroup C reveals the prehistoric migration routes of African exodus and early settlement in East Asia notes that “The phylogeographic distribution pattern of Hg C supports a single coastal ‘Out-of-Africa’ route by way of the Indian subcontinent, which eventually led to the early settlement of modern humans in mainland Southeast Asia” and concludes there was “a general south-to-north and east-to-west cline of Y-STR diversity is observed with the highest diversity in Southeast Asia.” The study Y-chromosome evidence of southern origin of the East Asian-specific haplogroup O3-M122 concludes O is also from the south. And last but not least, Hg D (your Japanese and Tibetan connection) is also deemed (Y chromosome evidence of earliest modern human settlement in East Asia and multiple origins of Tibetan and Japanese populations) to have come via the southern route although there are alternative theories and there may yet be surprises to come regarding this position. Furthermore, a northern origin for major Eurasian haplogroups M, N and R has been ruled out in this 2007 study. // One of the recent 2011 studies regarding the migration out of Africa routes “Extended Y chromosome investigation suggests postglacial migrations of modern humans into East Asia via the northern route” confirms a dominant southern migratory route but affirms a peopling of E. Asia also took place via a northern (Central-South Asian-cum-West European route:
“Our data, in combination with the published East Asian Y-haplogroup data, show that there are four dominant haplogroups (accounting for 92.87% of the East Asian Y chromosomes), O-M175, D-M174, C-M130 (not including C5-M356), and N-M231, in both southern and northern East Asian populations, which is consistent with the proposed southern route of modern human origin in East Asia. However, there are other haplogroups (6.79% in total) (E-SRY4064, C5-M356, G-M201, H-M69, I-M170, J-P209, L-M20, Q-M242, R-M207, and T-M70) detected primarily in northern East Asian populations and were identified as Central-South Asian and/or West Eurasian origin based on the phylogeographic analysis.
ANCIENT NORTH ASIAN = small eyes with single eye lid, sharp nose with nose bridge, triangle cheek and jaw bone (ugly eyes with pretty nose) === typical Korean, Mongolian, Ainu, AmerIndian….
ANCIENT CENTRAL ASIAN = eyes with double lid and muscle bag, round nose with no nose bridge, round cheek and jaw bone (pretty eyes) === typical Tibetan, Japanese, HakkaChinese…
ANCIENT SOUTH ASIAN = eyes with double lid, big nose === typical Vietnamese, Thai, Cantonese….
KOREAN = ancient HanChinese built Korean Dynasty, but Han could not outnumber the Native
JAPANESE = ancient HanChinese outnumber the Native, but Han adepted the Native language
TIBETAN= ancient HanChinese outnumber the Native, and still use similar ancient Han language (close to Hakka Chinese language)
CHINESE = ancient HanChinese were almost all killed by Mongolian in 13th century. Only modern Hakka Chinese is closer to Ancient HanChinese. Northern Chinese are closer to Mongol-Manchu, Southern Chinese are closer to South Asian.
Many japanese are born with monolid eyes :). And han chinese are southern chinese people.
Cantonese are not South Asian. Vietnamese is not all south Asians either. They originate in modern day China. Hakka, teochow, hakkien are all southern Chinese people. They are not northern Chinese at all. Not all northern people have sharp noses. The other day I saw a Korean lady from Moscow with the triangle jaw, cute nose, bu mono lid eyes, and very pretty skin. Her eyes aren’t ugly , but just different than southern chinese, thai, Vietnamese, etc. There are southern chinese with narrow noses and big eyes. I think southern chinese, Vietnamese, and thai people look much better than northern chinese, Koreans, and Japanese people. Many get plastic surgery to change their faces. What a shame. Hakka people do not all have round noses. Modern day chinese people are inter mixing with north and south. Look at Joan Chen (shanghainese) and her handsome husband is Cantonese (light skin, very tall sharp nose, and big eyes with double eyelids). Joan is not pretty at all. Flat nose, average eyes and not very appealing. She is very slender. In person she is very ugly than on camera.
Gee, I been mistaken as Japanese, Korean, Mexican, Thai, Philipina, Vietnamese, and bi-racial (Asian and white). I guess everyone has a sterotype of how other ethnic people looks like.
You can google DNA testing and send your sample through mail to them. Damn I hate all these hate. All asians came from one common ancestor and before that Asians came out of East Africa. Please stop the hate . Those that hate where taught to hate. Usually by their parents or relatives. Please judge people individual and not as a group. I learn that myself.
From a website . The truth with the exception of Indians because some are actually caucasian people just like the Iranians, etc originated from the same common ancestors. Modern day Europeans pretty much came from three branches of people. The dark skin color eyed people mixed with the light skin dark eye people along with Ancient Siberians. Just remember this people migrate from places to places, so what is Ancient Siberians may not be modern Siberians anymore. Siberians are Asian people. This fact and if you need the sicentic article about it just email me and I give you website. The origin of human beings are from East Africa. .We came from apes and some still deny it. What is Chinese, Korean and Japanese???? it is just a nationality but not an ethnicity. Southern Chinese don’t really look like Northern Chinese. Norther Chinese are usually very mongoloid looking like some Koreans and some Japanese. there is no country with such thing as pure blood.
I just wanted to know if it was tru that china exiled their people to the japan as a penal colonies for x prisoners ?
No, but population increases and influxes of migrants in Japan can be correlated to the many internal wars within China, i.e. suggesting refugees fleeing to Korea and Japan. In prehistoric to proto-historic era, prisoners and slaves were valuable currency, used as labour for building projects, megalithic tombs, the Great Wall, temples and palaces. Prisoners and slaves were often instead gifts and tribute sent to China.
You are talking a bullshit, My gene is Haplogroup M7 and I’m identified as a Jomon people who had arrived at Japan TWENTY THOUSANDS years ago, How comes your conclusion that M7 is in China? Do you mean to say that Japanese Jomon people are from China? Don’t be kidding
Please don’t be rude or I will remove all of your comments, unless you are willing to enter into a more open-minded and genial spirit of academic discussion.
M7 is well-researched by now, it is a pre-Jomon Upp Paleolithic mtDNA gene and is distributed in a widespread area over East Asia. A map of the distribution of M7, and the three main branches M7a, M7b and M7c can be seen here: http://nut.sakura.ne.jp/dokuwiki/doku.php?id=en:mtdna:m7
The original research study (from which the above map came) that first clarified and showed the M7 populations’ distribution (among other M populations) was the Kivisild study “The emerging limbs and twigs of the East Asian mtDNA tree” http://mbe.oxfordjournals.org/content/19/10/1737.full
This study was groundbreaking in that it identified and described new haplogroups M7, M8, M9, N9, and R9 and demonstrated the settlement processes of any particular region in appropriate time scale through the method of hierarchically subdividing the major branches of the mtDNA. That settlement process was illustrated by the characteristically southern distribution of haplogroup M7 in East Asia, whereas its daughter-groups, M7a and M7b2, specific for Japanese and Korean populations, testify to a presumably (pre-)Jomon contribution to the modern mtDNA pool of Japan.
Points to note about M7:
1. M7 seen over much of East Asia, see distribution map here
2. This page is a collection of linked articles, research that shows populations that carry the M7 lineages
3. M7 is very old, older than Jomon population. Japan is thought to be the terminus, end point of incoming M lineages. The older upstream M7 branches are thought to be in Cham and Viet populations in SEA. On age of M7, see Major Population Expansion of East Asians Began before Neolithic Time: Evidence of mtDNA Genomes, PLoS ONE 6(10): e25835. doi:10.1371/journal.pone.0025835 See Table 2 for the expansion times.
4. The origin of M7 is still debateable, though the general opinion is favour of a southerly (ie SEA) origin with the oldest detected branch in the Cham population. But M7 is also detected at lower frequencies among the Buryat, and the YAP gene trail leads through Central Asia, and M7 is known to have traveled together with Y-dna D, which leaves open the possibility of a more northerly entry point into Asia, via Central Asia. Alternatively, a western entry point into SEA is posited by some geneticists. It is considered that M7 splits off from M6. See Toomas and the M phylo tree at http://www.phylotree.org/tree/subtree_M.htm – see map at Toomas’s 2004 paper of this southern route/trail taken by macrohaplogroup M, and the broad areas from where the various haplogroups expanded, including M7 gene. Using this as a guide, plus various other miscellaneous papers on SEA (VN, Laos, JP and phylo gen tree of M).
How M7 splits off from M6:
Haplogroup M6 (Figure 6) is primarily found in the Indus Valley and on the western shores of the Bay of Bengal where its sub-clades M6a and M6b are concentrated towards the southwest and the northeast, respectively (Figure 1, panel M6, M6b cline is significant SAA p < 0.05, Figure 4). The highest frequencies of M6a and M6b were found amongst the Mukri scheduled caste from Karnataka (17%) and in Kashmir (10%), respectively. The Mukri form an endogamous group of no more than 10,000 individuals, who dwell on an area less than 2000 km2 [35]. That, together with the observation that all the sixteen M6 sequences found among the Mukri belong to a single haplotype, suggests that genetic drift has played a major role in the demographic history of the Mukri. The statistical significance of the high M6 frequency in Kashmir is undermined by the small sample size (19 individuals), which results in the very wide error margins for the frequency estimate (CR: 3.2–31.7%).
It might be helpful to keep an open mind about the origins of any one population as many studies keep coming onstream, and genotyping and gene distribution maps become more and more high resolution. In such, early pre-Jomon and even Jomon times when man were more primitive hunter-gathering small bands of people in East Asia, there were no political boundaries such as China, Korea and Japan, they freely came and went.
It might also be helpful to note, the Jomon people were already heterogenous in prehistoric times, i.e. they were of mixed lineages and therefore of mixed origins, and had different genetic combinations in different locations of Japan. See Adachi N., "Mitochondrial DNA analysis of the human skeleton of the initial Jomon phase excavated at the Yugura cave site, Nagano, Japan” See pdf full text
Abstract: In the present study, we performed a detailed mitochondrial DNA (mtDNA) analysis of a skeleton of the initial Jomon era unearthed from the Yugura cave site in Nagano, Japan, which was dated to 7920–7795 calBP by direct 14C dating. mtDNA of the Yugura skeleton was designated to haplogroup D4b2, which is widely observed in present-day East Asians, including the Japanese, but is absent in Hokkaido Jomon people. This finding indicates that the basal population of Japan was heterogeneous with respect to their mtDNA lineage. This is the first report on the genotype of the people from the initial phase of the Jomon period.
Yes you right ! Japanese came from jamon 20,000 years ago & Ainu too …Mongolia , native American Indians , & Maori are similar too
You beautify your word and claim that your conclusion is basic on the rearch,
What research did you do? Is that the research held your institution? Don’t be kidding, the link you gave me is totally from another website written by another people which is lack of reliability, Everyone can claim that his/her thinking is right,But this is just your GUESS, If following your theory,and all the Japanese people should had migrated from China,but the truth is that ONLY Yayoi people came from China. ALL the Jomon people are directly from southeast Asia where was priginally called Sundaland. And Japanese Haplogroup M7 has already arrived JAPAN before it appeared in China.If you want to demonstrate that your theory is right and I will ask you four questions.
1,What is the first mtDNA group coming to Japan?
2.You said that M7 group is divided into three subtypes,7a 7b 7c,right? What time did M7 started to separate? and how did they separated?
3. You gave me so many licks and complicated information?Are they all basic on the research of your institution? What is your institution? Unless I don’t know specifically of your institution for sure. What your clamied WILL NOT be trusted by publics. I watched many BBC documentaries hosted by Alice Robert who is a famous Paleontologist , but many people still question what she said about the Journey of human. She said that where people finally arrived is China but not Japan.actually this is totally WRONG.
I did not say that M7 came from China. I merely showed the distribution of M7 which is given by Toomas Kivilsd http://mega.bioanth.cam.ac.uk/ who is an authority on genetics. However, his map shows that many branches of M7 are also in China. The map merely shows that M7 likely originated somewhere in SEA (Cham populations) and separated into distinct regions M7a Japan; M7b Korea and M7c Philippines. And I am citing proper authorities as evidence in support of possible scenarios of how M7 arrived in Japan. The Sundaland theory is old, and somewhat superseded by many other theories. I also cite Adachi who is a Japanese scientist. Maybe you should update your reading, before you come on board. Many links are given as many pieces that fit as pieces of a puzzle. If you are not prepared to follow the evidence, please refrain from posting your own politically motivated drivel. If you have an alternative opinion, then state it with your own proper authorities, citations as evidence. Otherwise, you are wasting everybody’s time here.
I assume that I am not as eloquent as you are, because you are a SUCH A GREAT native English Scientist. One thing I want to say is that your theory offends many Japanese people including me, because we are directly from Sundaland, Sundaland theory is not old,and it is still prevalently used today on all sorts of services, Also Im here for justice, So I never feel regret of “wasting time ” like you said,Finally, I’m not the site owner , so I’m not fear of being deleted. And I have evidence, but I don’t want to show them here because this is only for someone I can trust.
😛 lol you do realize you look east asian. unless you have blue eyes and blonde hair, you can’t be offended when people called you a part of east asian.
Nobuyuki shinou is racist and he is like flat earthers that got offended because people give concrete proof that earth is not flat but a globe. i genuinely can’t believe there are some people actually think that earth is flat, but after seeing comment like nobutuki, that doesn’t seem so impossible.
thank you for wonderful and truthful answer @heritageofjapan. debunking myths, purist, mystery, etc 🙂
Interesting . So that’s why I look like japanese.
SMH at Nobuyuki….just give in to the fact that we are all just mutts.
Have you looked into Japanese and Hebrew genetic links?
Ashkenazi Jews, on top of their predominantly Near Eastern genes, have been found to have ancient East Asian mtDNAs from the A, N9a and M33 haplogroups see
“A genetic contribution from the Far East into Ashkenazi Jews via the ancient Silk Road”
https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4323646/
The above article puts the origins of M33 as being of Han Chinese of the Silk Route origins, but M33c may be found amongst M33c, whose subhaplogroups are very common among peoples like Japanese, Ainu, Koreans, Vietnamese, Tibetans, Nepalese, Bangladeshis, Siberians, and Asian Indian. Tey are also common among southern Han Chinese from Fujien province (but from other studies, we know these groups moved from more northerly psrts like North China or Southeast Asia).
Derenko’s study on rare northern and Eastern eurasian haplogroups (mtdna) has a section on N9a that identifies Japan as the most likely origin of N9a, and showed the expansion of N9a hgs on a cline from Eastern Eurasia all the way to Koros, Hungary. http://journals.plos.org/plosone/article?id=10.1371/journal.pone.0032179
The linked article below shows N9a shows up in Xiongnu ancient mtDNAs, so this suggests Xiongnu (synonymous with Hun traders) during the expansion of their Xiongnu nomadic empire likely would have had extensive contacts with Ashkenazi merchants along the Silk Route.
http://haplogroup.org/sources/mitochondrial-dna-diversity-in-a-transbaikalian-xiongnu-population/journals.plos.org/plosone/article?id=10.1371/journal.pone.0032179/
http://diabetes.diabetesjournals.org/content/56/2/518
This link shows A and N9a as dominant mtDNA hgs of the Japanese, however, hg A’s origins are harder to pinpoint, possibly originating in Korea. Haplogroup A has the most carriers in East Asia as well as its greatest degree of variety there so that is believed to be its point of origin. East Asians with varieties of A include Koreans, Tibetans, Vietnamese, Japanese, Ryukyuans, Han Chinese, Nakhi of China, Yi, Hmong, and several others. Varieties of A are also found in Siberians (Evenks, Buryats, Mansi, etc.), Mongols, Volga Tatars, Anatolian Turks, eastern Persians, Tajiks, Amerindians, and Inuit among others.
This is what humans did and still do: move, adapt, cultivate, fight, trade, and of course have lots kids.
Which us why we are all here right now, doing the same thing.
As a descendent of Asian peoples this doesn’t really surprise me. I’m glad the science is there to back it up. Thanks!
Those who can’t stand the idea that we’re all related, remember: you can chose your friends but you can’t chose your family 👨👩👦 (that’s everyone in the world!).
Pingback: Y-Chromosomes Suggest Origins of Japanese Bloodline - WARAUGUMI
I didn’t read it all, but I am making just a basic comment about what the Japanese actually are for anyone who stumbles onto this. This is backed by DNA evidence and migration logic used on other tested populations.
The Japanese are a mix of two people, Yayoi and Jomon. Jomon are a group which existed in Japan for tens of thousands of years, they originate from North Asia is hypothesised, as the only other groups which share the Y genes mutations as the Jomon are half of the Tibetan population and also Mongolian men.
A slight amount of it is found in Korea’s south also, but it’s small in number. Basically this old clan of Asiatic people originate from around Mongolia it is thought. They travelled a very long time ago before the more successful cultures along the Chinese Huang He evolved.
These people made it into Japan, the first people ever in Japan. They formed the Jomon culture, they also made it down into Tibet and scientists speculate that they also may have been more prevalent even in the Korea area and in China.
Though as you know other groups evolve and may become more dominant, it happens all the time. The birth of Chinese civilisation is along the Huang He and this group proved to be very successful, spreading all through Asia.
As they spread they took over, any areas where Jomon ancestors existed were pretty much on a genetic level taken over and at least on a Y mutation level are not detectable today in these areas.
Though places quite far away with more protection retained these genes, Mongolia is separated by a large mountain range and a desert from China, Japan is separated by sea and Tibet is on a giant Plateau so also was protected to some degree and hard to invade because of that.
The Yayoi finally did make it to Japan, these Yayoi certainly came across from the Korean peninsula and at least from skull remains they appear to be kind of like Koreans or Chinese people, yet have more slender faces for some reason.
This Yayoi more modern development of Asian basically interbred with the Jomon people in Japan who had been secluded in Japan for a very long time. Skulls and remains of the Jomon show wider eye sockets, wider heads and longer forearm bones, but also on average 1 inch shorter than the Yayoi people.
This formed the common Japanese person today, who I have read has on average around 22% Jomon mixture. Of course the people in Hokkaido have more Jomon on average as that was like the Jomon’s last refuge during the Yayoi takeover of Japan.
The Japanese in the Ryukyu Islands also have a high amount of Jomon DNA, as they also were much later met by the Yayoi as they were secluded by water.
So it’s quite clear the Jomon did actually inhabit all of Japan, but the Yayoi first arrived in the southern mainland of Japan (where Jomon DNA is the lowest today, so it makes total sense)
___________________________________________________________
And this last part is my estimate, this is what I believe are the average percentages based on the overall DNA average when adjusted for the Y mutation occurrence difference per region.
South Koreans = 3% Jomon DNA on average.
Southern mainland Japan = 15% Jomon DNA on average.
Central mainland Japan = 18% Jomon DNA on average.
Northern mainland Japan = 23% Jomon DNA on average.
Hokkaido (northern Island) = 37% Jomon DNA on average.
Ryukyu Japan (far southern Islands) = 31% Jomon DNA on average.
These numbers are adjusted for population size per region also. Working out an average of around 22% Jomon DNA over the entire country.
If we are to consider the distribution and frequency of the paternal haplogroup O1b2 (previously O2b), than it is most likely that the ancestors of the yayoi people were from Korea. However, it is clear to me that the Yayoi and modern koreans are not the same. Heres why:
Japan has a lack of haplogroup C2 which was previously known as C3. A very recent study that analyzed dna extracted from cigarettes of north korean origin revealed that the total frequency of C2 (20.3%) rivaled that of O1b2 (24.4%). Infact, a previous sampling that analyzed the y haplogroups of koreans in southwestern manchuria revealed that C2 (23%) was higher than O1b2 (19.5). Even in South Koreas coastal regions haplogroup C2 can appear significantly say 18 to 20 percent. If you ignore the jomon component of modern japanese and focus on the yayoi part it is clear that O1b2 is by far the most dominant. However, in modern Korea thats not the case because in koreans O2 which was previously known as O3 is the dominant clade. Now its pretty clear to myself that yayoi is not the same with modern koreans. The question is why?
That is a relatively easy question to answer. C3 (y-chromosome) represents the descendants of Genghis Khan (and possibly his progenitor ancestor) https://www.ncbi.nlm.nih.gov/pubmed/22452430
North Korea, China and tos ome extent South Korea were overrun by Genghis Khan’s desdendant Mongols who never reached Japan which is consistent with Japanese historical accounts.
The korean haplogroup C2 clades are not the same as those in Mongolia. Mongols are vastly C2b while koreans are mostly C2c sublineage. Its not just Koreans but all east asian populations have only a slight frequency of C2b. In koreans it ranges from 1 to 6.9 percent. Its quite surprising that in the north korean data only a single cigarette out of a total of 123 belonged to C2b while the other clades were C2c. Infact, a tincy bit of C1a1 was detected. On the other hand, mongolians and central asians posses only a bit of what is known as C2c.
Please help me locate my lost great grandfather Shomitshu, Japanese soldier who fought in Indonesia and married my grandmother who is from an island of Indonesia.
Pingback: 【セルビア】DMM英会話【ボスニア】 - 英会話学習・勉強法2chまとめ
Hi, is this site still being serviced? Do you have links to any studies showing differing levels of HLA for Japanese versus Chinese/Korean, etc.?
(yes, we are still here) There is data showing subsets shared between differing regions and Japan. Findings from the paper Detection of Ancestry Informative HLA Alleles Confirms the Admixed Origins of Japanese Population https://journals.plos.org/plosone/article?id=10.1371/journal.pone.0060793 “We hypothesized that the LD across HLA alleles characterized by low frequency in Okinawa (CL3 and CL4) are strong because HLA haplotypes carrying these alleles had been recently derived from the Korean Peninsula and expanded in Japan’s mainland rapidly. Thus, we examined genetic relationship between Japanese and South Korean. We compared the frequencies of haplotypes carrying A*33:03 allele that was characterized by high frequency in South Korean (Table 4). There were four common haplotypes carrying A*33:03 in South Korean. While the haplotype A*33:03-B*44:03-C*14:03-DRB1*13:02 was frequent in mainland Japanese and South Korean, the other three haplotypes were frequent in South Korean but rare or absent in Japanese. By searching the AFND database, the haplotype A*33:03-B*44:03-C*14:03-DRB1*13:02 was observed only in Japanese and Korean. In contrast, the other three haplotypes were prevalent in East and Southeast Asian populations (Table 4) [21], [33]–[36]. The haplotype A*24:02-C*12:02-B*52:01-DRB1*15:02 was observed only in Japanese and South Korean. These findings may reinforce our hypothesis that the origin of H1 and H2 haplotypes was the Korean Peninsula.” (see https://journals.plos.org/plosone/article?id=10.1371/journal.pone.0060793 for HLA allele chart comparisons between regions)
See also by the same lead author of all the papers: Distribution of HLA haplotypes across Japanese
Archipelago https://www.nature.com/articles/jhg201590.pdf?origin=ppub and The Admixed Origin of Japanese Population from HLA Alleles https://www.jstage.jst.go.jp/article/mhc/21/1/21_37/_article
Notwithstanding ever intense focus on Korean contribution to Japanese origins however, this month’s published paper makes it clear that there is a third dominant contribution (Han Chinese) to the Kofun era population and to Japanese origins, and it would be remiss of me not to mention this paper “Ancient genomics reveals tripartite origins of Japanese populations” https://www.science.org/doi/10.1126/sciadv.abh2419?_ga=2.20572152.1927151092.1631694601-1492079976.1608110151