Note that C and Z are descended from M8 which is also present in Japan.
According to an earlier study by Tanaka Masashi’s “Mitochondrial Genome Variation in Eastern Asia and the Peopling of Japan”
“With less than 14 conglomerates, the Japanese, including Ainu and Ryukyuans, were part of a big group formed by Korean, Buryat, Tibetans, and northern Chinese. Ainu was the first differentiated Japanese sample. Ryukyuans separated later, when mainland Japanese and Koreans still comprised a single group. The lack of homogeneity between Ainu and Ryukyuans was pointed out by Horai et al. (1996), who questioned that they shared a recent common ancestor. The main differences between them were attributed to two dominant clusters (C1 and C16, … respectively) present in Ainu but absent in Ryukuyans, and two Ryukyuan dominant clusters (C3 and C13, … respectively) absent in Ainu.” Also …
“A monophyletic clade (Fig. 1A) groups M8a, C, and Z lineages. Mutations 4715, 15487T, and 16298 have been proposed as diagnostic for this clade (Yao et al. 2002a). The transversion 7196A and the transition 8584 should also be included in its definition (Fig. 1A; Kivisild et al. 2002). However, as the 248d is also shared by all Z and C lineages (Fig. 1A), a basal node defined by this deletion and named CZ has been recently proposed (Kong et al. 2003). Subhaplogroup C was RFLP-defined by Torroni et al. (1992) by +13262 AluI. Yao et al. (2002a) added 248d, 14318, and 16327 as characteristic of C. In addition, positions 3552A, 9545, and 11914 are also diagnostic of this clade (Fig. 1A; Kivisild et al. 2002). The Japanese TC52 has the C1 status and the Buryat 6970 and the Evenky 6979 have the C4 status proposed by Kong et al. (2003). Subhaplogroup Z was defined by Schurr et al. (1999) by the presence of the following noncoding motifs: 16185, 16223, 16224, 16260, and 16298. Recently, it was considered that only 16185 and 16260 mutations should be counted as basic for the group (Yao et al. 2002a). However, in full agreement with the characterization proposed on the basis of complete Chinese Z sequences (Kong et al. 2003), three additional mutations (6752, 9090, and 15784) have been placed on the basal branch of Z (Fig. 1A). We detected four Japanese Z clades that, in addition, shared mutation 152 and another without it. Tentatively, they have been named from Z1 to Z5 (Fig. 1A). Yao et al. (2002a) defined M8a by 14470, 16184, and 16319 transitions. Two more mutations (6179 and 8684) are also characteristic of this subhaplogroup (Kong et al. 2003). In Japanese we have found that 16184 is not harbored by all M8a members. Consequently, lineages with this mutation have M8a2 status and those lacking it M8a1 status (Fig. 1A). The largest diversities for C are in Korea (100%), central Asia (86%), and northern China (78%-74%). Therefore, C can be considered a clade with a Northeast Asian radiation. Representatives of subhaplogroup Z extend from the Saami (Finnilä et al. 2001) and Russians (Malyarchuk and Derenko 2001) of west Eurasia to the people of the eastern peninsula of Kamchatka (Schurr et al. 1999). Its largest diversities are found in Koreans (88%), northern China (73%), and central Asia (67%), compatible with a central-East Asian origin of radiation for this group. Finally, M8a has its highest diversity in Koreans (100%), and southern (100%) and eastern Chinese, including Taiwanese (73%). Thus, southeastern China was a potential focus of radiation of this group. All these subhaplogroups are present in mainland Japanese but neither in Ryukyuans nor in Ainu.”
Abstract Summary
The human mitochondrial haplogroup C1 has a broad global distribution but is extremely rare in Europe today. Recent ancient DNA evidence has demonstrated its presence in European Mesolithic individuals. Three individuals from the 7,500 year old Mesolithic site of Yuzhnyy Oleni Ostrov, Western Russia, could be assigned to haplogroup C1 based on mitochondrial hypervariable region I sequences. However, hypervariable region I data alone could not provide enough resolution to establish the phylogenetic relationship of these Mesolithic haplotypes with haplogroup C1 mitochondrial DNA sequences found today in populations of Europe, Asia and the Americas. In order to obtain high-resolution data and shed light on the origin of this European Mesolithic C1 haplotype, we target-enriched and sequenced the complete mitochondrial genome of one Yuzhnyy Oleni Ostrov C1 individual. The updated phylogeny of C1 haplogroups indicated that the Yuzhnyy Oleni Ostrov haplotype represents a new distinct clade, provisionally coined “C1f”. We show that all three C1 carriers of Yuzhnyy Oleni Ostrov belong to this clade. No haplotype closely related to the C1f sequence could be found in the large current database of ancient and present-day mitochondrial genomes. Hence, we have discovered past human mitochondrial diversity that has not been observed in modern-day populations so far. The lack of positive matches in modern populations may be explained by under-sampling of rare modern C1 carriers or by demographic processes, population extinction or replacement, that may have impacted on populations of Northeast Europe since prehistoric times.

Network representation of C1 HVR-I sequences in Mesolithic Yuzhnyy Oleni Ostrov and modern Eurasian populations.
Each haplotype is represented by a circle, the area of which is proportional to the number of individuals that were found to carry this haplotype in the literature. The haplotypes are colour-coded according to their geographical location: India (black), Asia (dark grey), Lebanon (light grey), and Europe (white). Each section of the circles represents individuals sampled from a same population. Mutations are all substitutions and are reported according to the Reconstructed Sapiens Reference Sequence minus 16000. The star represents the hypervariable region-I haplotype that characterizes the root of the C1 clade [possible origin in the Amur River region]. The haplotype labeled ‘UZOO’ is the hypervariable region-I haplotype sequenced from individuals of the archaeological site of Yuzhnyy Oleni Ostrov. All the other haplotypes were found in modern populations.
Human mitochondrial haplogroup (hg) C is part of the non-African macro-haplogroup M. Most of the diversity of hg C is found today in indigenous populations of Asia and the Americas [1]–[2]. In northern Asia, hg C represents, together with hg D, more than half of the present-day mitochondrial (mtDNA) diversity [3]. Haplogroup Z, the sister-clade of hg C, has a broad distribution ranging from northern Scandinavia (in Saami) to central Asia, Siberia, northern China and Korea.
Phylogenetic analyses of complete mtDNA genomes revealed four major sub-clades of hg C, termed C1, C4, C5 and C7 (e.g., [3]–[7]). Of these, haplogroup C1 has one of the broadest distributions of all human mtDNA hgs in the world, ranging from Iceland to East Asia and the Americas. The C1 basal haplotype is defined by the hypervariable region I and II (HVR-I and HVR-II) motif: A16129G, T16187C, C16189T, G16230A, T16278C, T16298C, C16311T, T16325C, C16327T (HVR-I; numbering according to the Reconstructed Sapiens Reference Sequence RSRS; [8]) and C146T, C152T, C195T, A247G, A249d, 290–291d and T489C (HVR-II).
The phylogeny of hg C1 is structured into five distinct monophyletic sub-clades, C1a, C1b, C1c, C1d and C1e, which exhibit a clear geographical distribution pattern ([4], [7], [9]–[10]; Figure 1). Three of the C1 sub-clades (C1b, C1c and C1d) are restricted to Native American populations, although spread widely across the American continent [11]–[12]. It was proposed that these three Native American C1 sub-clades were among the ancestral founder lineages, along with hg A2, B2 and D1, which reached the Americas during the initial human colonisation of the continent [4]–[5], [7], [9]. The source population of this migration was assumed to be in eastern Asia, where most of the diversity of hg C is observed today, and where C1a, a sister clade of the American C1 clades, is found at low frequencies in diverse indigenous populations [9]. The peopling of the Americas was made possible by the Beringian ice-free land bridge that connected north-east Siberia and Alaska before (∼30,000 years Before Present, BP) and after (∼13,000 yBP) the last Ice Age [9], [13]. The place of origin of ancestral hg C1 was approximated in the Amur River region just south of Beringia (eastern Asia) on the basis of the current frequency distribution of hg C1 in Asia [9]. The last hg C1 clade to have been described, C1e, was only found recently in a few individuals in Iceland and was shown to be distinct from any of the previously defined Asian and American clades on the basis of seven coding region and three control region mutations [10].
In Europe, the dense and extensive sampling of the HVR-I diversity has revealed extremely low frequencies of hg C1, with very few haplotypes found in Germans [14], Canarians [15], Icelanders[16]–[17] and Bashkirs [18] (Figure 2). These sequences lack HVR-I Single Nucleotide Polymorphisms (SNPs) diagnostic of the sub-clades C1a (T16356C) and C1d (A16051G). However, a more detailed assignment of the European haplotypes into sub-haplogroups is limited by the low resolution provided by HVR-I and the lack of information from the coding region thus far. These limitations therefore impede the reconstruction of their precise phylogenetic placement, origin and relation to the Asian, American and Icelandic sister-clades.
Three hypotheses for the origins of the C1 lineages in Europe can be put forward [10], [16]–[17]. Hypothesis 1 proposes a recent genetic input from Asia into Europe during historical times. Historically, Central and East Europe experienced repeated influences from invading groups from the neighbouring Asian steppes, which could have introduced C1 into Europe. Well-documented examples include the Huns from Mongolia in the 4th–5th centuries Anno Domini (A.D.) and the Mongols in the 13th century A.D.[19]. However, the common Asian C1a clade is characterised by the HVR-I transition T16356C, which has not been found in any European C1 haplotype. In the case of a recent Asian ancestry, a reversal of the mutation at nucleotide position (np) 16356 in all the European sequences would be required. Hypothesis 2 assumes an American origin, where hg C1 would have reached Europe through admixture between Native Americans and Europeans. This gene flow may have occurred during and after the colonization of the New World by Europeans in the 15th century A.D., i.e. in post-Columbian times [16]–[17]. Alternatively, one explanation for the presence of the sub-clade C1e in Iceland was a pre-Columbian admixture between Native Americans and Icelandic Vikings, which are widely acknowledged to have built temporary pioneer settlements in the north-western coast of the Americas in the 10th century A.D. [10]. In accordance with the hypothesis of the American origin, few European C1 HVR-I sequences could belong to either the C1b or C1c American clades, as diagnostic SNPs for these two clades are located outside HVR-I [16]–[17]. Hypothesis 3, proposes that hg C1 has been present in Europe since prehistoric times in the light of the recent finding of hg C1 HVR-I haplotypes in three individuals of the 7,500-year-old Mesolithic site of Yuzhnyy Oleni Ostrov (individuals UZOO-7, UZOO-8, and UZOO-74), North West Russia (Figure 1;[20]). The classification of the corresponding mtDNA haplotype within hg C1 was previously determined by HVR-I sequencing (hg C1 defining mutation T16325C) as well as by typing informative SNPs in the coding region (hg C defining mutation A13263G [20]), but the lack of resolution of the HVR-I sequence prevented establishing clear phylogenetic relationships with currently known hg C1 clades.
In this study, we sequenced the complete mtDNA genome of one of the three Mesolithic hg C1 carriers from the Yuzhnyy Oleni Ostrov archaeological site (individual UZOO-74) in order to shed further light on the population history of the Yuzhnyy Oleni Ostrov hunter-gatherers and to contribute to the characterisation of the mtDNA diversity, evolutionary history and phylogeography within hg C1.
To this day, complete mtDNA genome sequences from ancient specimens have successfully been determined for a Palaeo-Eskimo from Greenland [21], the 5,000-year-old Tyrolean Iceman [22], a 700-year-old individual from New Zealand [23], a Palaeolithic individual from Tianyan, China [24], as well as several Palaeolithic, Mesolithic and Neolithic individuals from Europe [25]–[28]. Most of these mtDNA genomes were obtained on high-throughput ‘next-generation’ sequencing platforms (e.g., [29]). In accordance with these recent studies, we first created a genomic library, which was subsequently enriched for mtDNA in two iterative rounds of hybridisation to in-house designed biotinylated DNA probes, following the protocol by [28]. The enriched DNA library was sequenced on an Ion Torrent PGM platform. We analysed the resulting mtDNA genome from the Yuzhnyy Oleni Ostrov specimen in the light of an updated phylogeny of all currently available hg C1 lineages. The resulting mtDNA genome sequence allowed us to identify a novel C1 sub-clade, coined “C1f”, which fills a gap in the knowledge of the hg C1 distribution in West Eurasia.
Ancient Mitochondrial Genome Sequencing
Our ancient DNA (aDNA) enrichment, followed by sequencing on an Ion Torrent PGM platform, allowed the unambiguous determination of 99.8% (16537 out of 16569 base pairs, bp) of the UZOO-74 mtDNA genome with 20,579 unique reads assembled to the RSRS at an average coverage of 68X (Table 1) and average read length of 55±14.5 bp. Indels, a well-defined homopolymer sequencing error, were observed in the resulting data set. However, adequate depth and coverage of the mtDNA genome sequence data prevented false-positive base calls.
Missing data comprised 32 consecutive bp, spanning nps 7525–7556 (Table 1). Other mtDNA genome sequences that we have generated following the same protocol have also exhibited a low coverage or dropout in exactly the same region [28]. Figure S1 shows that low GC content regions are characterised by a poor coverage; in particular in the region 7525–7556, GC content is only 25.0% compared to 44.4% GC for the whole mtDNA. It is therefore suspected that this region of the mtDNA genome is energetically sub-optimal (AT-rich) for two rounds of hybridisation and stringency washes in the ionic and temperature conditions used here [28], which may produce secondary DNA structures that adversely affect hybridisation-based DNA capture [30]. In addition, re-amplification of the enriched libraries was done with AmpliTaq Gold (Applied Biosystems; see [28]), a Taq polymerase known to be biased towards high GC content [31].
The ancient mtDNA haplotype of individual UZOO-74 differed from the RSRS at 58 nucleotide positions (Table 1). Among these, 51 substitutions define the sub-hg C1 in accordance with the current phylogeny (www.PhyloTree.org), including back mutations at T182C! and G11914A!, which are identical by state to the RSRS. In addition, UZOO-74 showed five private substitutions (G247A!, A8577G, A11605t, A12217G and T16189C!, including two additional back mutations in the hypervariable region). These five additional nucleotide differences were directly amplified and sequenced from two different extracts in order to verify whether they represented true private mutations defining a novel C1 sub-clade (Table S1). All five mutations have been confirmed by direct sequencing and were taken into account in further phylogenetic analysis of hg C1 sequences. Importantly, we did not observe any SNPs characteristic of other hgs, nor any mixed signals that could indicate systematic DNA degradation or DNA contamination from exogenous sources.
We analysed the pattern of nucleotide misincorporation at the 3′ and 5′-ends of the DNA fragments in order to assess whether the estimated age of the molecules reflects the age of the sample [32]–[33]. We observed a C-to-T substitution frequency of 22.4% at the 5′-end (Figure S2), which sits well with previous findings that suggested a correlation between this frequency and the age of the samples [34], whereby samples older than 500 years had a C-to-T substitution frequency >10%. The Bayesian statistical framework implemented in mapDamage v2.0.1 [33] also provided simulated posterior distribution of three parameters of the damage model: λ, probability of terminating in overhang; ∂D, probability of cytosine deamination in double strands; and ∂S, probability of cytosine deamination in single strands. The posterior distribution of these parameters all departed from 0 (λ: mean, 0.582; standard deviation, 0.021; ∂D: mean, 0.036; standard deviation, 0.001; ∂S: mean, 0.651; standard deviation, 0.041), in accordance with the DNA sequences generated from the UZOO-74 individual arising from aDNA molecules, and not from contamination by more recent DNA molecules during post-excavation handling. The results of the DNA damage analyses support the authenticity of the aDNA data presented here.
The Sequence from Mesolithic Yuzhnyy Oleni Ostrov Defines a Novel Lineage within the C1 Phylogeny
Upon confirmation of the five novel mutations in the C1 mtDNA genome of individual UZOO-74, we genotyped the same SNPs in the two other C1 individuals UZOO-7 and UZOO-8 from Yuzhnyy Oleni Ostrov. Direct sequencing confirmed the presence of all five novel SNPs, suggesting that the three C1 individuals from Yuzhnyy Oleni Ostrov were maternally related. Their mtDNA genomes may be strictly identical, or they may display differences in the form of additional private SNPs at coding region positions that have not been sequenced in these remaining individuals. The precise nature of the genetic relationships between these individuals cannot be inferred from the archaeological and genetic data currently available.
A search against the public Phylotree database yielded no match for the newly sequenced C1 mtDNA haplotype in 16810 modern complete mtDNA genomes (entries, mtDNA tree build 15 (30 Sep 2012) on PhyloTree.org [35]). The five SNPs identified in these individuals, and among these, the three coding region mutations A8577G, A11605t and A12217G in particular, represent novel sub-clade defining mutations that have not been reported together within a single hg C1 haplotype before. We therefore assigned them to a distinct new clade, which we tentatively named “C1f” following the conventional nomenclature (Figure 3). The resulting phylogenetic reconstruction shows that clade C1 is now characterised by six monophyletic sub-clades: C1a, C1b, C1c, C1d, C1e and C1f. The tree topology suggests that the Eurasian C1 sub-clades, the East Asian C1a, the rare C1f branch from Yuzhnyy Oleni Ostrov and the Icelandic C1e split early from the most recent common ancestor of the C1 clades and evolved independently (Figure 3).
DISCUSSIOn
Under-sampling of the Mitochondrial Genome Diversity
In the present study, we established that the hg C1 mtDNA genome sequence carried by the Mesolithic individuals of the Yuzhnyy Oleni Ostrov site in north-western Russia defines a new clade, C1f, within the hg C1 phylogeny. Because of the polytomous topology of the hg C1 tree, no direct phylogenetic relationship could be established between C1f and the other well geographically defined C1 clades. As a result, clear inferences regarding the origin and evolutionary history of the C1f clade will remain difficult to draw, unless future sequencing of complete mtDNA genomes uncovers sequences closely related to the C1f genome sequenced here.
The absence of a direct match with sequences in databases of complete mtDNA genomes could be explained by under-sampling of mtDNA genomes in modern human populations. The number of published modern-day Homo sapiens complete mtDNA genome sequences is still small compared to that of HVR-I sequences. As such it is not too surprising that studies regularly report the discovery of novel clades and lineages (e.g., hg C1e [10]; within hg C1d [7]). Furthermore, the geographical coverage of modern-day populations for complete mtDNA genome sequencing is still unequally distributed, and the sampling so far has focused either on few specific populations or on particular hgs (e.g., [36]–[38]). As a consequence, mtDNA genomes available from the literature can still only provide an incomplete yet biased picture of the full, extant mtDNA diversity.
Absence of Match for C1f in Asia
Asia, and more precisely Siberia, could be considered as potential places of origin for the C1f clade identified in the Mesolithic site of Yuzhnyy Oleni Ostrov. This hunter-gatherer group was indeed shown to exhibit mtDNA affinities with modern-day populations of western and southern Siberia, the Altai region, or Mongolia [20]. The hypothesis of an Asian origin for the C1f sub-clade is also supported by the fact that most of the diversity of hg C is found in present-day populations of East Eurasia [3]. Sequences closely related to hg C1f may persist in modern-day populations of East Eurasia but remain undetected to date, as mtDNA genomes for these populations have not been as densely and extensively sampled as, for example, European populations.
Absence of Match for C1f in Europe
Despite the dense sampling of mtDNA in modern-day populations of Europe, only a few hg C1 HVR-I and no hg C1f mtDNA genome sequences were detected. The close matches for the HVR-I sequence of C1f did not display the back mutation T16189C! (Figure 2) and hence, none matched the C1f HVR-I haplotype exactly. However, np 16189 has been described as one of the top five transitional hotspots in the human control region [39], and hence provides little phylogenetic discrimination power. It is possible that these European haplotypes belong to the C1f clade without harbouring the mutation at np 16189. Therefore, additional SNPs in the coding region are required to definitely rule out these Eurasian C1 haplotypes as potential members of the C1f clade, and potential persistence of hg C1f in Europe since the Mesolithic.
Extinction or Near-extinction of C1f due to Post-Mesolithic Population Dynamics
Low frequencies and a restricted distribution seem to have been characteristic of hg C1 already in Mesolithic times, as hg C1 could not be detected in any of the other European Mesolithic populations sampled for ancient mtDNA in Eurasia further west [20]: in central/eastern Europe [25], [27], [40], and in Scandinavia [41]. This suggests an under-sampling of Mesolithic populations for aDNA, mating isolation of the Yuzhnyy Oleni Ostrov population, and/or influences from Siberian populations that had not reached Central Europe. Because of its low frequency, the distribution of hg C1 is prone to be affected by demographic processes, such as genetic drift or population replacements that may have occurred since Mesolithic times. Eventually, hg C1 may have reached extremely low frequencies or have gone extinct, thus preventing it from being detected in present-day European populations. The effects of these population processes can be observed at the population level, as the Yuzhnyy Oleni Ostrov group, similarly to the other described Mesolithic populations of Europe, was indeed shown to exhibit little genetic continuity with present-day Europeans [20]. Significant dissimilarities have been shown between the mtDNA gene pool of European Mesolithic populations characterised by a low diversity and high frequencies of hg U sub-clades (U2, U4, U5 and U8 in particular), and the rather homogeneous mtDNA makeup of present-day Europeans, which arrived during the Neolithic transition and subsequent periods [20], [40]–[42].
Absence of Match for C1f in the Americas
The Americas also remain under-sampled for complete mtDNA genomes and could be suggested as a potential geographical origin for the C1f lineage, as it has been for the Iceland-restricted C1e sub-clade[10]. For C1e, an American origin through mating of Viking explorers with Native American women sometime earlier than 300 years ago was proposed by [10]. Among other hypotheses including that of a European origin, an American origin was favoured on the basis that most of the hg C1 diversity is found on the American continent, despite the fact that no sequence belonging to hg C1e could be detected in the Americas (or anywhere else). This lack of match was explained by under-sampling of the American mtDNA genome diversity [10]. In any case, if admixture between Native Americans and Vikings did occur, it must have been limited, as no other American-specific lineage (e.g. hg A2, B2, D1, C1b, C1c, C1d) was detected in Iceland.
As for Mesolithic Europe, the possibility of a direct prehistoric genetic influence from the Americas is highly unlikely. However, in the eventuality that further sampling of complete mtDNA genomes in the Americas reveals the presence of additional haplotypes belonging to C1f, it would suggest an evolutionary history similar to that of mtDNA hg X2. Like hg C1, hg X2 displays relatively low frequencies albeit with a global distribution in the Northern hemisphere. For example, clade X2a was observed in Europe in the West, in the Near East, Europe, Central Asia, Siberia as well as North America [43]. One model for the present-day distribution of hg X2 suggests that clade X2a split early from the rest of the X2 lineages in the Near East, and reached east Siberia before participating in the second wave of migration into the Americas through admixture with Beringian populations [44]. A similar scenario involving an early split of the different C1 clades in Asia followed by their spread and subsequently isolated evolution could be considered as an explanation for the wide geographical distribution of hg C1 in general. However, this scenario currently lacks substantial support.
Similar Genetic Pre-history for the Icelandic-specific C1e and the Mesolithic C1f European Sub-clades
While the updated phylogeography of hg C1 does not allow defining the precise origins and divergence times of the C1f and C1e clades, the observation of C1f in Mesolithic Yuzhnyy Oleni Ostrov brings us to reconsider the hypotheses concerning the origins of C1e. Building on a hypothesis proposed by [10], we suggest that the Icelandic-specific C1e sub-clade could have had a recent origin in northern Europe rather than an American origin. This hypothesis is relevant with regard to the origins of the Icelandic population, as Iceland was discovered and first settled by Scandinavian Vikings around 1,130 years ago. Vikings raids extended as far from their homeland in Scandinavia as France, Spain and Sicily, but their main expansion range comprised western Russia, the Baltic region, Scandinavia, and the British Isles [16]. The study of the mtDNA diversity of present-day Icelanders identified that most of the Icelandic mtDNA lineages had Norse (from Scandinavia) or Gaelic origins (from the British Isles) and that the Icelandic gene pool had strongly been impacted by genetic drift [16]–[17], [45].
Considering the Scandinavian origins of Icelanders and the identification of the sister clade C1f in Mesolithic North East Europe, it can be proposed that the Icelandic-specific C1e and C1f sub-clades might have both split from the common ancestors of the C1 lineages somewhere in Eurasia and later reached northern Europe during independent or similar migrations (before the Mesolithic for C1f). Therefore, the rare occurrence of the C1e and C1f sub-clades in Europe could be the result of their dilution within the pre-existing European mtDNA diversity when these lineages reached Europe. Of note, a contrasting pattern of elevated frequency and diversity was observed for the American C1 sister-clades (C1b, C1c and C1d): all three American sub-clades signal important population expansion during the initial peopling of the continent, which was void of human occupation and thus competing lineages. The distribution of the C1e sub-clade restricted to Iceland, associated with the presence of the novel sub-clade C1f in a region neighbouring the homeland of Vikings and clearly predating the Viking expansion, lends support to the hypothesis that hg C1e might have been brought in by the Vikings who first colonised Iceland. The presence of a novel sub-clade (C1f) closely related to the Icelandic-specific C1e sub-clade in a region neighbouring the homeland of Vikings and clearly predating the Viking expansion lends support to the hypothesis that hg C1e might have been brought in by the Vikings who first colonised Iceland. While the C1e sub-clade might have been preserved at detectable frequencies in the Icelandic population due to the effects of founder event, it most likely has gone extinct in the source population in northern Europe as a consequence of its low frequency. In contrast, due to the small size of the population through time, Icelandic mtDNA diversity has been greatly affected by genetic drift and increased rates of mtDNA haplotype extinctions [45]. As such, the C1e clade would be more likely to survive in the potential North European source population than in Iceland [45], but the extensive sampling of the Icelandic population makes it more likely to be detected there than anywhere else in North Europe. The potential long-term survival of C1 lineages in prehistoric Europe is highly relevant to the discussion about the prehistoric interactions between the ancestral populations of Europeans, Siberians and Native Americans. It is consistent with recently published genomic data from a 24,000 year-old Upper Paleolithic individual from Mal’ta, South Siberia [46]. Interestingly, this individual was shown to belong to the western Eurasian hg U, which was also the most frequent hg found in Yuzhnyy Oleni Ostrov Mesolithic individuals (64%) [20]. Genome-wide data from Upper Palaeolithic Mal’ta revealed affinities with both present-day western Eurasian and Native Americans, and further supports gene-flow between the ancestral populations of Europeans and Native Americans prior to the colonisation of the Americas [46]. The new C1f lineage thus bridges the geographic gap between the Icelandic, the Siberian and the Native American C1 lineages and argues for the presence of C1 lineages, albeit at low frequency, in prehistoric West Eurasia
Related studies:
Clio Der Sarkissian, et al., Ancient DNA Reveals Prehistoric Gene-Flow from Siberia in the Complex Human Population History of North East Europe
Abstract Summary
Abstract
North East Europe harbors a high diversity of cultures and languages, suggesting a complex genetic history. Archaeological, anthropological, and genetic research has revealed a series of influences from Western and Eastern Eurasia in the past. While genetic data from modern-day populations is commonly used to make inferences about their origins and past migrations, ancient DNA provides a powerful test of such hypotheses by giving a snapshot of the past genetic diversity. In order to better understand the dynamics that have shaped the gene pool of North East Europeans, we generated and analyzed 34 mitochondrial genotypes from the skeletal remains of three archaeological sites in northwest Russia. These sites were dated to the Mesolithic and the Early Metal Age (7,500 and 3,500 uncalibrated years Before Present). We applied a suite of population genetic analyses (principal component analysis, genetic distance mapping, haplotype sharing analyses) and compared past demographic models through coalescent simulations using Bayesian Serial SimCoal and Approximate Bayesian Computation. Comparisons of genetic data from ancient and modern-day populations revealed significant changes in the mitochondrial makeup of North East Europeans through time. Mesolithic foragers showed high frequencies and diversity of haplogroups U (U2e, U4, U5a), a pattern observed previously in European hunter-gatherers from Iberia to Scandinavia. In contrast, the presence of mitochondrial DNA haplogroups C, D, and Z in Early Metal Age individuals suggested discontinuity with Mesolithic hunter-gatherers and genetic influx from central/eastern Siberia. We identified remarkable genetic dissimilarities between prehistoric and modern-day North East Europeans/Saami, which suggests an important role of post-Mesolithic migrations from Western Europe and subsequent population replacement/extinctions.
This work demonstrates how ancient DNA can improve our understanding of human population movements across Eurasia.
It contributes to the description of the spatio-temporal distribution of mitochondrial diversity and will be of significance for future reconstructions of the history of Europeans.
Figure 1. Map of Eurasia showing the approximate location of ancient (uncalibrated dates) and present-day Eurasian samples.
Red dots represent the archaeological sites sampled for ancient mitochondrial DNA in this study: aUZ, Yuzhnyy Oleni Ostrov; aPo, Popovo; aBOO, Bol’shoy Oleni Ostrov. Black circles represent ancient populations abbreviated as follows: aEG, Confederated nomads of the Xiongnu (2,200–2,300 yBP); aKAZ, Nomads from Kazakhstan (2,100–3,400 yBP); aKOS, Kostenski individual (30,000 yBP); aKUR, Siberian Kurgans (1,600–3,800 yBP); aLOK, Lokomotiv Kitoi Neolithic individuals (6,130–7,140 yBP); aPWC, Scandinavian Pitted-Ware Culture foragers (4,500–5,300 yBP); aUST, Ust’Ida Neolithic population (4,000–5,800 yBP). Smaller black dots signify the location of Palaeolithic/Mesolithic sites sampled for ancient mitochondrial DNA in aHG (4,250–15,400 yBP). Present-day populations are abbreviated as follows: alt, Altaians; BA, Bashkirs; BU, Buryats; CU, Chuvash; EST, Estonians; FIN, Finns; ket, Kets; kham, Khamnigans; khan, Khants; KK, Khakhassians; KO, Komis; KR, Karelians; LTU, Lithuanians; LVA, Latvians; man, Mansi; ME, Mari; MO, Mordvinians; MNG, Mongolians; NEN, Nenets; nga, Nganasans; NOR, Norwegians; tof, Tofalars; tuv, Tuvinians; UD, Udmurts; SA, Yakuts; saa, Saami; sel, Selkups; SWE, Swedes. The approximate location of the Volga-Ural Basin and of the different regions of Russian Siberia are also indicated.
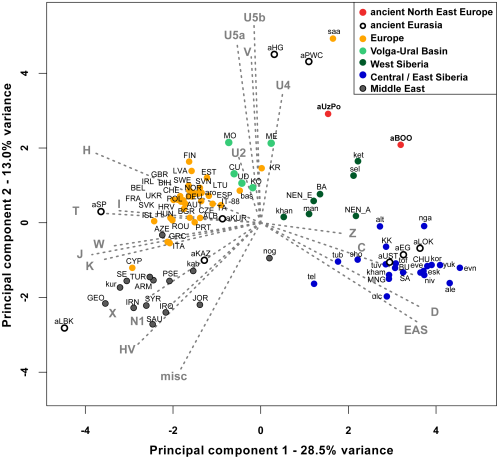
FIGURE 2: Principal Component Analysis of mitochondrial haplogroup frequencies.
The first two dimensions account for 41.5% of the total variance. Grey arrows represent haplogroup loading vectors, i.e., the contribution of each haplogroup. Red dots represent ancient populations described in this study: aUzPo, Yuzhnyy Oleni Ostrov and Popovo (7,500 uncal. yBP); aBOO, Bol’shoy Oleni Ostrov (3,500 uncal. yBP). Other ancient populations were labeled as follows: aEG, Confederated nomads of the Xiongnu (4,250-2,300 yBP); aHG, Palaeolithic/Mesolithic hunter-gatherers of Central/East Europe (4,250-30,000 yBP); aKAZ, Nomads from Kazakhstan (2,100–3,400 yBP); aKUR, Siberian Kurgans (1,600–3,800 yBP); aLBK, Neolithic individuals from Germany (7,000–7,500 yBP); aLOK, Lokomotiv Kitoi Neolithic individuals (6,130–7,140 yBP); aSP, Neolithic individuals from Spain (5,000–5,500 yBP); aPWC, Scandinavian Pitted-Ware Culture foragers (4,500–5,300 yBP); aUST, Ust’Ida Neolithic population (4,000–5,800 yBP). Extant populations were abbreviated as follows: ALB, Albanians; ale, Aleuts; alt, Altaians; ARM, Armenians; aro, Arorums; AUT, Austrians; AZE, Azerbaijani; BA, Bashkirs; bas, Basques; BEL, Belarusians; BGR, Bulgarians; BIH, Bosnians; BU, Buryats; CHE, Swiss; CHU, Chukchi; CU, Chuvashes; CYP, Cypriots; CZE, Czechs; DEU, Germans; esk, Eskimos; ESP, Spanish; EST, Estonians; eve, Evenks; evn, Evens; FIN, Finns; FRA, French; GBR, British; GEO, Georgians; GRC, Greeks; HRV, Croatians; HUN, Hungarians; ing, Ingrians; IRL, Irish; IRN, Iranians; IRQ, Iraqi; ISL, Icelanders; IT-88, Sardinians; ITA, Italians; JOR, Jordanians; kab, Kabardians; ket, Kets; kham, Khamnigans; khan, Khants; KK, Khakhassians; KO, Komi; kor, Koryaks; KR, Karelians; kur, Kurds; LTU, Lithuanians; LVA, Latvians; man, Mansi; ME, Mari; MNG, Mongolians; MO, Mordvinians; NEN_A, eastern Nenets; NEN_E, western Nenets; nga, Nganasans; niv, Nivkhs; nog, Nogays; NOR, Norwegians; POL, Poles; PRT, Portuguese; PSE, Palestinans; ROU, Romanians; RUS, Russians; SA, Yakuts; saa, Saami; SAU, Saudi Arabians; SE, Ossets; sel, Selkups; sho, Shors; SVK, Slovakians; SVN, Slovenians; SWE, Swedes; SYR, Syrians; TA, Tatars; tel, Telenghits; tof, Tofalars; tub, Tubalars; TUR, Turks; tuv, Tuvinians; UD, Udmurts; UKR, Ukrainians; ulc, Ulchi; vep, Vepses; yuk, Yukaghirs.
doi:10.1371/journal.pgen.1003296.g002
Relevant excerpts:
“… populations of the ‘Central/East Siberian’ cluster were predominantly composed of hgs A, B, C, D, F, G, Y, and Z, while in contrast populations of the ‘European’ cluster were characterized by higher frequencies of hgs H, HV, V, U, K, J, T, W, X, and I (e.g., [43–47]). The two ancient groups – aUzPo and aBOO – from two individual time periods appeared remarkably distinct on the basis of the PCA, suggesting a major genetic discontinuity in space and time.
Comparison of Mesolithic Yuzhnyy Oleni Ostrov/Popovo (aUzPo) with extant populations of Eurasia The hg distribution in the Mesolithic aUzPo population: U4 (37%), C (27%), U2e (18%), U5a (9%), and H (9%), indicated an ‘admixed’ composition of ‘European’ (U4, U2e, U5a and H, 73%) and ‘Central/East Siberian’ (C, 27%) hgs, based on the PCA plot (Figure 2). Interestingly, the population of aUzPo did not group with modern NEE populations, including Saami, but fell instead between the present-day ‘European’ and ‘Central/East Siberian’ clusters on the PCA graph, and more precisely between populations of the VUB (in light green) and West Siberia (in dark green). The high frequency of hg U4 is a feature shared between Mesolithic aUzPo, present-day VUB (Komi, Chuvashes, Mari), and West Siberian populations (Kets, Selkups, Mansi, Khants, Nenets), with the latter group also being characterized, like aUzPo, by the presence of hg C. The genetic affinity between Mesolithic aUzPo and present-day West Siberian populations could be visualized on the genetic distance map of North Eurasia (Figure 3A), on which locally lighter colorings indicated low values of genetic distances, and therefore an affinity between aUzPo and extant West Siberians.
In order to test the potential population affinities formulated on the basis of the hg-frequency PCA and the distance map, we examined the present-day geographical distribution of the haplotypes found in aUzPo via haplotype sharing analyses (Figure 4). These analyses are less impacted by biases due to small population sizes or unidentified maternal relationships in ancient populations, and thus are less prone to artefacts. Although the highest percentages of shared haplotypes for aUzPo were observed in pools of West Siberian Khants/Mansi/Nenets/Selkups (2.8%), South Siberian Altaians/Khakhassians/Shors/Tofalars (2.2%) and Urals populations (Chuvash/Bashkirs, 2.0%), matches were widely distributed across Eurasia. … the area of maximam similarity for aUzPo lay in West Siberia. …
While the Mesolithic aUzPo site showed genetic affinities with extant populations of West Siberia in hg-based analyses, the precise genetic origins of aUzPo individuals was more difficult to identify from haplotypic data due to the high number of basal haplotypes. At the archaeological level also, the Siberian connection with aUzPo is less clear. The material culture present in the burials of aUzlinks these populations with the neighboring regions in the West but also in the East and Sout-East … As for Siberia, it has undergone a complicated early and mid-Holocene migration history due to repeated environmental changes… With the data at hand, it is therefore difficult to make any definite statement about sixth millennium connections between Karelia and Siberia.
Interesting, samples from aBOO, which are 4,000 years younger and located further North-West than aUzPo were characterized by a large proportion and elevated diversity of mtDNA lineages showing a clear ‘Central/East Siberian’ orgin (hgs C, D, And Z). Haplogroups C and D are the most common hgs in northern, central and eastern Asia. They are thought to have originated in eastern Asia and expanded through multiple migrations after the Late Glacial Maximum (~20,000 yBP … Notably, haplotypic matches were observed between aBOO and modern-day Siberian Buryats of the peri-aikal region, which was proposed to be the origin of ancient migrations that dissemminated hgs C and D… Today, the sharp western boundary for the distribution of hgs C, D and Z lies in the VUB, (0.2-0.9%), nad D (0.9-12%)… Sub-hgs Z1 n D5 are also present in modern-day Saami, with highest cumulated frequencies (15.9%) in the Saami of Finland, the easternmost part of the Saami geographical distribution… A precise date for the arrival of these ‘ Central/East Siberian’ lineages in the 3,500 year-old aBOO site indicates that an eastern genetic influence pre-dates historical westward expansions from Central/East Siberia of , e.g., the Huns and the Mongols (~400-1,500 AD). We present here direct genetic evidence for a prehistoric gene-flow from Siberia. On the basis of modern-day genetic data, hg Z1 was proposed to have been introduced into populations of the VUB and Saami by migrations from Siberia via the southern Urals to the Pechora and Vychegda basins of northwest Urals, associated with the appearance of the Kama culture ~8,000 yBP … The presence of hg Z1 in a BOO establishes a direct genetic link between aBOO and modern-day population of the VUB and Saami, and possibly indicates the trajectory of the migration that brought ‘Central/East Siberian’ lineages into NEE. The fact that aBOO did not contain any other Saami-specific haplotypes suggests an independent origin and contribution of Z1 to the Saami gene pool.
The genetic links between the sample populations of aUzPo/ABOO and the extant populations of Siberia follow a general pattern discussed for the early and mid-Holocene (6000-10000 yBP). Facilitated by the East–West extension of vegetation zones between the Russian Far East and Eastern Europe long-distance contacts and connections across Eurasia have been proposed for a number of cases. For example, the North East and East European hunter-gatherer pottery is thought to have originated in the early ceramic traditions of the Russian Far East and Siberia… An eastern Asian origin followed by a westward expansion was also discussed for domesticated broomcorn millet (Panicum miliaceum L. …”
“The major prehistoric migration in this area was associated with the spread of early pottery from the East into the Baltic, Karelia and Fennoscandia starting around 7,000 yBP. This migration might have contributed to an early population change in Karelia and Fennoscandia as well, but the mtDNA characteristics of the populations involved is presently unknown [76-78]. As for Siberia a general push-back of populations by an expansion of populations from the South-West is discussed… Thus, the present-day distribution of populations similar to aUzPo and aBOO might just be a remnant of a once much larger extension across western and Central northern Eurasia, which is consistent with frequencies of hgs U4 and U5 ie the Palaeolithic/Mesolithic genetic substratum, have remained higher in extant pplations of NEE, the VUB and Western Siberia than in central Europeans, where these were largely replaced at the onset of the Neolithic … Genetic discontinuity between aUzPo, aBOO and present-day populations of NEE was also observed at the haplotype level as seen by the lack of matches between lineages from ancient individuals and from present-day NEE (eg ‘Central/ East Siberian’ lineages in a BOO), or by their total absence in all Eurasian populations of the comparative dataset. A good example is the haplotype C1 found in aUzPo, which is absent in modern–day Eurasians and in allother foraging populations of Europe. This indicates that hg C1 was rare and probably preserved in a UZPo by a relative reproductive isolation, previously proposed for Mesolithic hunter-gatherers of NEE on the basis of odontometric and craniometric analyzes. These results do not exclude a common origin for European foragers but highlight differentiating consequences of post-glacial founder effects followed by reproductive isolation among Palaeoithic/Mesolithic groups. Genetic discontinuity between prehistoric populations of Europe may have been caused by the random loss of genetic diversity through drift, which is likely to have been accelerated in small and isolated groups, such as aUzPo and aBOO. In the Kola Peninsula, the scarcity in the archaeological records observed in the Kola Peninsula for the Early Metal Age was interpreted as an indication of drastic size reductions of human groups, as a response to deteriorating climatic conditions ~2,500 yBP .. This could have lead to the local extinction of mtDNA lineages of Siberian origin detected in aBOO in the Kola Peninsula.”
…
Derenko M., et al., PLoS ONE 5(12): e15214. doi:10.1371/journal.pone.0015214 Origin and post-glacial dispersal of mitochondrial DNA haplogroups C and D in northern Asia
More than a half of the northern Asian pool of human mitochondrial DNA (mtDNA) is fragmented into a number of subclades of haplogroups C and D, two of the most frequent haplogroups throughout northern, eastern, central Asia and America. While there has been considerable recent progress in studying mitochondrial variation in eastern Asia and America at the complete genome resolution, little comparable data is available for regions such as southern Siberia–the area where most of northern Asian haplogroups, including C and D, likely diversified. This gap in our knowledge causes a serious barrier for progress in understanding the demographic pre-history of northern Eurasia in general. Here we describe the phylogeography of haplogroups C and D in the populations of northern and eastern Asia. We have analyzed 770 samples from haplogroups C and D (174 and 596, respectively) at high resolution, including 182 novel complete mtDNA sequences representing haplogroups C and D (83 and 99, respectively). The present-day variation of haplogroups C and D suggests that these mtDNA clades expanded before the Last Glacial Maximum (LGM), with their oldest lineages being present in the eastern Asia. Unlike in eastern Asia, most of the northern Asian variants of haplogroups C and D began the expansion after the LGM, thus pointing to post-glacial re-colonization of northern Asia. Our results show that both haplogroups were involved in migrations, from eastern Asia and southern Siberia to eastern and northeastern Europe, likely during the middle Holocene.
…
More than a half of the northern Asian pool of mtDNA is fragmented into a number of subclades of haplogroups C and D, two of the most frequent haplogroups throughout northern, eastern, central Asia and America. Previous studies have proposed that haplogroups C and D originated around 30–50 kya in eastern Asia, from where they subsequently expanded northwards to southern Siberia, and further deep into northern Asia and the Americas, and westwards along the Steppe Belt extending from Manchuria to Europe [14], [15]. It has been also shown that haplogroups C and D were strongly involved in the late-glacial expansions from southern China to northeastern India [16]. In addition, because of their high frequency and wide distribution, haplogroups C and D most likely participated in all subsequent episodes of putative gene flow in northern Eurasia. These include (i) the Paleolithic colonization of Siberia that is associated with the development of macroblade industries (40–30 kya), (ii) further recolonization and possible replacement of early Siberians by microblade-making human populations from the Lake Baikal, Yenisei River, and Lena River basin regions (20 kya), (iii) appearance of pottery-making Neolithic tradition in the forest-steppe belt of northern Eurasia starting at about 14.5 kya and its expanding into the East European Plane (7 kya), (iv) the Neolithic dispersal of agriculture in eastern Asia, (v) the expansion of the Afanasievo and Andronovo cultures (5–3 kya), and (vi) more recent events of gene flow to eastern and central Europe.
As a result, it is likely that the dissection of haplogroups C and D into subhaplogroups of younger age and more limited geographic and ethnic distributions might reveal previously unidentified spatial frequency patterns, which in turn could be correlated to prehistoric and historical migratory events. However, until now, haplogroups C and D have been resolved genealogically only partially allowing for the identification of seven principal subclades (C1, C4, C5, C7, D4, D5, D6) and some of their internal subclades, the phylogeography of which has been evaluated only in some instances [4], [5], [9], [10], [12], [13], [16]–[18].
To shed some light on the origin and dispersal of haplogroups C and D in Asia, we present here an analysis of the complete mtDNA genomes from populations distributed over the geographical range of these two haplogroups.
Results and Discussion
The spread of haplogroups C and D
Haplogroups C and D display an extremely wide geographic distribution and high frequencies over most of their range. Haplogroup C peaks over 50% among Yukaghirs of northeastern Asia, central Siberian Yakuts and Evenks as well as East-Sayan Tofalars. Its frequency is persistently above 20% in Altaian, West-Sayan and Baikal region populations and drops to 13% or less among Chukchis, Eskimos and Itelmens in the east, Altaian Kazakhs, Shors, and Oroks in the south, and Khants and Kets in the west. The diminishing line (frequencies under 5%) goes through the Turkic and Finno-Ugric populations of the Volga basin, further south through the populations of the Caucasus and western Asia. In the southern direction the decline of haplogroup C frequency is almost as sharp as in the west direction: it is very common in Mongolia (15%) and most of the populations of central Asia (7–18%), but occurs as rarely as 1–5% in Korea, China, Thailand, Japan, Island southeastern Asia and India. Haplogroup C is detected at a very low frequency in several populations of eastern and central Europe and virtually absent in western Europe and Africa (Table S1).
The second most common haplogroup in all northern Asian populations is haplogroup D, which is also very common in eastern, central Asia and America. Haplogroup D encompasses almost 20% of the total mtDNA variation in most of northern Asia and retains a very high overall frequency in all regional northern Asian groups (11–34%), central Asian (14–20%) and eastern Asian (10–43%) populations (Table S2). Its frequency declines towards the west and south, to 2% or less in India and western Asia, but in the Caucasus, Volga-Ural Region and southeastern Asia is still as high as 5–10%. Interestingly, haplogroup D is also found in some northeastern Europeans, like Karelians, Saami and Scandinavians, while haplogroup C is absent among them (Table S2).
The phylogeny of haplogroup C
The phylogeny of the C sequences is illustrated in Figure S1. The average sequence divergence of the 174 C complete genome corresponds to a coalescence time estimate of 27.37 (19.55; 35.44) kya when using the sequence variation of the entire genome and 26.33±6.58 when only synonymous mutations are considered [19] (Table S3). The C tree shows an initial deep split into four sister subclades, C1, C4, C5 and C7, each containing several independent basal branches, one within C1, at least three within C4, four within C5, and three within C7 (Figure S1). The C1 branch is represented by C1a subclade which is a sister clade of the Native American subclades C1b, C1c, and C1d, which are dated to 18.6±2.3 kya [5], [9] and most likely arose early – either in Beringia or at a very initial stage of the Paleoindian southward migration [4]. The Asian C1a-branch derived likely from the same ancestral population as the three Native American subclades [4] shows a relatively lower coalescence time varying from 2 to 8.5 kya (1.97±1.97 kya for synonymous clock rate and 8.57 (2.6; 14.75) kya for complete mtDNA clock rate), implying that its expansion from Beringia occurred long after the end of the LGM.
The C4 branch shows a coalescence time of 20–22 kya, implying that it began to expand before the LGM. Inside haplogroup C4 a new subclade, C4e, specific for Altai region populations has been revealed (Figure S1). It is defined by transitions at nps 151, 152, 7307, 15479 and, together with Russian individual (Rus_184), characterized by lack of adenine insertion at np 2232, which is thought to be diagnostic for a whole subclade C4a’b’c [20]. This subclade represents a major fraction of C4 mtDNAs and can be further subdivided into C4a, C4b and Native American-specific branch C4c identified so far only in two Ijka-speakers from Colombia [4] and one Shuswap individual from British Columbia [21]. Cluster C4a dates to 19–25 kya, demonstrating the pre-LGM time of divergence, in contrast to C4b, which is characterized by younger coalescence time estimated as 6–7 kya.
The other major branch of the tree, C5 has a coalescence time of 14–17 kya, depending on the mutation rate used. The phylogeny of haplogroup C5 reveals at least four subhaplogroups (C5a-C5d) with the similar coalescence time estimates varying from 9 to 14 kya (Table S3). The C7 branch is the most ancient, with an estimated coalescence time of 26–28 kya, but in contrast to C4 and C5, which encompass the entire geographical range of C, C7 is present mainly in eastern Asian and northeastern Indian populations (Figure S1).
Based on complete mtDNA genome sequence information, we have identified several new subclusters within the C4 (C4a1a1, C4a1a2, C4a1a2b, C4b4, C4b5, C4b6, C4b8) and C5 (C5a2a1, C5b, C5b1a, C5c1, C5c2) subclades, as well as redefined some previously described clusters. Complete mtDNA sequence based phylogeographic analysis has shown a remarkable geographic distribution for some of haplogroup C subclusters (Figure 1). Thus, certain subclades of C4 and C5 were more prevalent in the southern Siberian populations being found mainly in Altai-Sayan and Baikal region populations (C4a1a1, C4a1a2b, C4b4, C4b5, C4b6), whereas others (C4b2, C4b7, C4b8, C5a2a) were found only in Arctic populations of Chukchi, Koryaks, Nganasans, and Yukaghirs. Interestingly, subclusters C4a1b, C4a2a2a, C4a2b, C4a2a2, and C7a1a encompass predominantly Indian mtDNA genomes, and show evolutionary ages within time frames of 8–20.5 kya. It is worth emphasizing that the ages of northern Asian clusters fall into the ranges of 3–14.5 kya, whereas the coalescence time estimates for Arctic region-specific lineages are not exceed 4.5 kya.
Figure 1. Complete mtDNA phylogenetic tree of haplogroup C.
This schematic tree is based on phylogenetic tree presented in Figure S1. Time estimates (in kya) shown for mtDNA subclusters are based on the complete mtDNA genome clock (the first value) and the synonymous clock (the second value) [19]. The size of each circle is proportional to the number of individuals sharing the corresponding haplotype, with the smallest size corresponding to one individual. Geographic origin is indicated by different colors: northeastern Asian – in blue, central and southern Siberian – in green, eastern Asian – in red, Indian – in grey, European – in yellow, and others (i.e.of unknown population origin) – in white.
doi:10.1371/journal.pone.0015214.g001
Four of the new and two previously published sequences (one Teleut and one Tubalar from the Altai region of southern Siberia, three Poles from northern Poland, and one FamilyTreeDNA project individual of unknown ancestry) clustered into uncommon branch, named C5c, harboring the diagnostic motif 10454-16093-16518T-16527. Several mtDNAs with the same control-region motif were detected earlier at a low frequency in some European, Asian and southern Siberian populations – in Poles (0.4%), Belorussians (0.3%), Romanians (0.6%), Persians (0.2%), Kirghiz (1.1%), Altaians (0.9%), Teleuts (7.5%), Khakassians (0.9%) and Shors (4%) [4], [10], [22]–[28]. With the exception of mtDNAs from southern Siberia, which harbored additional control region transition at np 16291, all other C5c mtDNAs were characterized by another control region mutation at np 16234. The complete mtDNA genome phylogeny confirms that the C5c branch shows an initial split into two sister subclades, one encompassing mtDNAs from Europe (C5c1) and the other consisting of only two sequences from the Altai region of southern Siberia (C5c2) (Figure 2). It appears that European branch C5c1 is more differentiated, as far as two of three sequenced Polish mtDNAs formed a separate branch (C5c1a), defined by a coding region mutation at np 7694. The relatively large amount of internal variation accumulated in the Polish branch of C5c would mean that C5c1 arose in situ in Europe after the arrival of a C5c1 founder mtDNA from southern Siberia, and that C5c1 affiliation is a marker of maternal Siberian ancestry. The phylogeny depicted in Figure S1 provides additional information concerning the entry time of the founder mtDNA – the age of C5c node is estimated as 9.7 (3.17; 16.49) kya when using the sequence variation of the entire genome, and 9.2±4.74 when only synonymous mutations are considered (Table S3). The early presence of mtDNA lineages of eastern Asian ancestry in Europe is further confirmed by the discovery of a N9a haplotype in a Neolithic skeleton from the Szarvas site, located in southeastern Hungary that belonged to the Körös Culture, which appeared in eastern Hungary in the early 8th millennium B.P. [29]. …
… It should be noted that dispersal of Saami-specific Z1a mtDNAs shared a common ancestry with lineages from the Volga-Ural region as recently as ~3 kya probably chronicles the same expansion [32].
Conclusions
The peopling of northern Asia by anatomically modern humans probably began more than 40 kya, with the first evidence in the Altai region, suggesting the southern mountain belt of Siberia and Middle Siberian plateau as a likely route for this pioneer settlement of northern Asia[33]–[36]. The present-day variation of haplogroups C and D suggests that these mtDNA clades had already expanded before the LGM, with their oldest lineages being present in the eastern Asia. In particular, most of the eastern Asian subclades of haplogroup D show coalescence ages of between 15 and 42 kya, thus suggesting that some of them were already present here before the LGM. As for northern Asia, most of the present-day southern and northeastern Siberian variants of haplogroups C and D started to expand after the LGM. This can be partially ascribed, as in Europe [31], [37]–[39] and southeastern Asia [40], to the (re)colonization processes of areas which were unsuitable for human occupation during the LGM due to aridity and lower temperatures. The Late Glacial re-expansion of microblade-making populations from the small refugial areas in southern Yenisei and Transbaikal region of southern Siberia at the end of the Ice Age from ~18 kya could be suggested as a major demographic process signaled in the mtDNA by the distribution of northern Asian-specific subclades of haplogroups C and D. The age of haplogroup C5, ~14–17 kya, supports this postulated arrival after the LGM, as does the age of the D2 and D4b1a2, which date to ~11–21 kya. However, all northeastern Asian-specific subclades present ages lower than 10 kya, so it is possible that their arrival into the Arctic region of northern Asia occurred later, in Holocene.
Importantly, we have not found in northern Asia any genetic signatures of sufficient antiquity to indicate traces of pre-LGM expansions, that originated from the Upper Paleolithic industries that were present both in the southern Siberia and Siberian Arctic, and that date back to ~30 kya, well before the LGM [1], [34], [36]. Apparently, the Upper Paleolithic population of northern Asia did not leaving a genetic mark on the female lineages of modern Siberians. It is probable that the initial population expansion in the southern Siberia region involved maternal lineages other than haplogroups C and D. Nevertheless none of the remaining northern Asian haplogroups became as frequent in Siberia as haplogroups C and D.